-Search query
-Search result
Showing 1 - 50 of 16,051 items for (author: yu & g)
EMDB-42603: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/hexamer)
EMDB-42625: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC804515)
EMDB-42626: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/up)
EMDB-42627: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/down)
EMDB-44748: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer)

PDB-8uv2: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/hexamer)

PDB-8uvo: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC804515)

PDB-8uvp: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/up)

PDB-8uvq: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/down)

PDB-9boq: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer)

EMDB-41766: 
CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant, scFv16, and dopamine

EMDB-41776: 
CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant and dopamine

PDB-8tzq: 
CryoEM structure of D2 dopamine receptor in complex with GoA KE Mutant, scFv16, and dopamine

PDB-8u02: 
CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant and dopamine

EMDB-44551: 
Map of eastern equine encephalitis virus q3 spike protein in complex with VLDLR without masked refinement
EMDB-44255: 
Cryo-EM structure of the mouse TRPM8 channel in the ligand-free desensitized state

EMDB-44256: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014

EMDB-44257: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMG2850
EMDB-44258: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMTB
EMDB-44259: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 and the cooling agonist C3
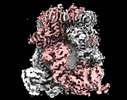
EMDB-44260: 
Cryo-EM structure of the avian great tit TRPM8 channel in complex with the antagonist TC-I 2014

EMDB-44261: 
Cryo-EM structure of the mouse TRPM8 channel in complex with PI(4,5)P2 and Ca2+
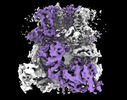
EMDB-44262: 
Cryo-EM structure of the mouse TRPM8 channel in complex with Ca2+ in the absence of PI(4,5)P2

PDB-9b6d: 
Cryo-EM structure of the mouse TRPM8 channel in the ligand-free desensitized state

PDB-9b6e: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014

PDB-9b6f: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMG2850

PDB-9b6g: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMTB

PDB-9b6h: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 and the cooling agonist C3

PDB-9b6i: 
Cryo-EM structure of the avian great tit TRPM8 channel in complex with the antagonist TC-I 2014

PDB-9b6j: 
Cryo-EM structure of the mouse TRPM8 channel in complex with PI(4,5)P2 and Ca2+

PDB-9b6k: 
Cryo-EM structure of the mouse TRPM8 channel in complex with Ca2+ in the absence of PI(4,5)P2
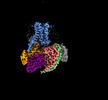
EMDB-60628: 
Carazolol-activated human beta3 adrenergic receptor
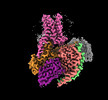
EMDB-60629: 
Epinephrine-activated human beta3 adrenergic receptor

PDB-9ijd: 
Carazolol-activated human beta3 adrenergic receptor

PDB-9ije: 
Epinephrine-activated human beta3 adrenergic receptor

EMDB-31367: 
Structure of mumps virus nucleoprotein without C-arm

PDB-7exa: 
Structure of mumps virus nucleoprotein without C-arm

EMDB-50124: 
Mammalian ternary complex of a translating 80S ribosome, NAC and NatA/E

EMDB-50125: 
Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E

EMDB-50126: 
Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E-HYPK

EMDB-42487: 
Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer of dimers

EMDB-38850: 
Cryo-EM structure of human dopamine transporter in apo state

EMDB-38851: 
Cryo-EM structure of human dopamine transporter in complex with dopamine

EMDB-38852: 
Cryo-EM structure of human dopamine transporter in complex with benztropine

EMDB-38853: 
structure of a proteinACryo-EM structure of human dopamine transporter in complex with GBR12909

EMDB-38854: 
Cryo-EM structure of human dopamine transporter in complex with methylphenidate

PDB-8y2c: 
Cryo-EM structure of human dopamine transporter in apo state

PDB-8y2d: 
Cryo-EM structure of human dopamine transporter in complex with dopamine

PDB-8y2e: 
Cryo-EM structure of human dopamine transporter in complex with benztropine

PDB-8y2f: 
Cryo-EM structure of human dopamine transporter in complex with GBR12909
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
