-Search query
-Search result
Showing 1 - 50 of 1,103 items for (author: scott & h)

EMDB-43514: 
SPOT-RASTR - a cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface

PDB-8vt0: 
SPOT-RASTR - a cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface

EMDB-41908: 
Local refinement map on VFT-CRD of active-state CaSR in lipid nanodiscs

EMDB-41909: 
Consensus refinement map of active-state human CaSR in lipid nanodiscs

EMDB-41910: 
Local refinement map on CRD-7TM of active-state CaSR in lipid nanodiscs

EMDB-41925: 
Local refinement map on VFT-CRD of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs

EMDB-41926: 
Local refinement map on Gi of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs

EMDB-41927: 
Local refinement map on CRD-7TM of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs

EMDB-41928: 
Consensus refinement map of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs

EMDB-41949: 
Consensus refinement map of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs

EMDB-41950: 
Local refinement map on CRD-7TM of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs

EMDB-41951: 
Local refinement map on VFT-CRD of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs

EMDB-41952: 
Local refinement map on Gq of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs

EMDB-41953: 
Consensus refinement map of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs

EMDB-41954: 
Local refinement map on VFT-CRD of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs

EMDB-41956: 
Local refinement map on CRD-7TM of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs

EMDB-41957: 
Local refinement map on Gi of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs

EMDB-29560: 
5-MeO-DMT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex

EMDB-29571: 
4-F, 5-MeO-PyrT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex

EMDB-29585: 
Vilazodone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex

EMDB-29597: 
LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex

EMDB-29599: 
Buspirone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex

PDB-8fy8: 
5-MeO-DMT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex

PDB-8fye: 
4-F, 5-MeO-PyrT-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex

PDB-8fyl: 
Vilazodone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex

PDB-8fyt: 
LSD-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex

PDB-8fyx: 
Buspirone-bound serotonin 1A (5-HT1A) receptor-Gi1 protein complex
EMDB-43683: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound
EMDB-43684: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound

PDB-8vzn: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound

PDB-8vzo: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound
EMDB-40180: 
MsbA bound to cerastecin C

PDB-8gk7: 
MsbA bound to cerastecin C

EMDB-19856: 
Focused map 1- K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-19857: 
Focused map 2 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-19858: 
Focused map 3 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-19859: 
Focused map 4 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-19860: 
Focused map 5 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide

EMDB-41363: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5

PDB-8tl6: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5

EMDB-29951: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#1 of 20)

EMDB-29952: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#2 of 20)

EMDB-29953: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#3 of 20)

EMDB-29954: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#4 of 20)

EMDB-29955: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#5 of 20)
EMDB-29956: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#6 of 20)
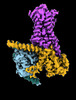
EMDB-29958: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#7 of 20)
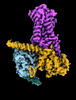
EMDB-29959: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#8 of 20)
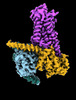
EMDB-29960: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#9 of 20)
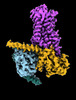
EMDB-29961: 
CryoEM structure of beta-2-adrenergic receptor in complex with nucleotide-free Gs heterotrimer (#10 of 20)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
