-Search query
-Search result
Showing 1 - 50 of 121 items for (author: rima & l)

EMDB-17757: 
Cryo-EM structure of the Cas12m-crRNA-target DNA complex

PDB-8pm4: 
Cryo-EM structure of the Cas12m-crRNA-target DNA complex
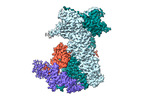
EMDB-18498: 
Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex

PDB-8qmo: 
Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

PDB-8fxb: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

PDB-8fxc: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

PDB-8s9g: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment

EMDB-26855: 
Arabidopsis DDM1 bound to nucleosome (H2A.W, H2B, H3.3, H4, with 147 bp DNA)

PDB-7ux9: 
Arabidopsis DDM1 bound to nucleosome (H2A.W, H2B, H3.3, H4, with 147 bp DNA)

EMDB-28758: 
Calcitonin Receptor in complex with Gs and Pramlintide analogue peptide San45

EMDB-28759: 
Human Amylin3 Receptor in complex with Gs and Pramlintide analogue peptide San385

EMDB-28810: 
Human Amylin3 Receptor in complex with Gs and Pramlintide analogue peptide San385 (Cluster 5 conformation)

EMDB-28812: 
Amylin 3 Receptor in complex with Gs and Pramlintide analogue peptide San45

PDB-8f0j: 
Calcitonin Receptor in complex with Gs and Pramlintide analogue peptide San45

PDB-8f0k: 
Human Amylin3 Receptor in complex with Gs and Pramlintide analogue peptide San385

PDB-8f2a: 
Human Amylin3 Receptor in complex with Gs and Pramlintide analogue peptide San385 (Cluster 5 conformation)

PDB-8f2b: 
Amylin 3 Receptor in complex with Gs and Pramlintide analogue peptide San45

EMDB-29930: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1

PDB-8gcc: 
T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1
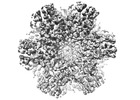
EMDB-16187: 
Wild tye immature Gag Structure

EMDB-16190: 
KAKA mutation immature Gag structure

EMDB-29790: 
30S focus refined map of WT E.coli ribosome complexed with A-site ortho-aminobenzoic acid charged tRNA-Phe

EMDB-29786: 
Structure of WT E.coli 70S ribosome complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site ortho-aminobenzoic acid charged NH-tRNAPhe

EMDB-29788: 
Structure of WT E.coli ribosome 50S subunit with complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site 3-aminopyridine-4-carboxylic acid charged NH-tRNAPhe

PDB-8g6w: 
Structure of WT E.coli 70S ribosome complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site ortho-aminobenzoic acid charged NH-tRNAPhe

PDB-8g6y: 
Structure of WT E.coli ribosome 50S subunit with complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site 3-aminopyridine-4-carboxylic acid charged NH-tRNAPhe

EMDB-29787: 
Structure of WT E.coli ribosome 50S subunit with complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site meta-aminobenzoic acid charged NH-tRNAPhe

PDB-8g6x: 
Structure of WT E.coli ribosome 50S subunit with complexed with mRNA, P-site fMet-NH-tRNAfMet and A-site meta-aminobenzoic acid charged NH-tRNAPhe

EMDB-28092: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-093

EMDB-28090: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-040

EMDB-28091: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-045
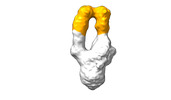
EMDB-28093: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-156
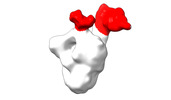
EMDB-28094: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-234

EMDB-28095: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-260

EMDB-28096: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-279

EMDB-28097: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-290
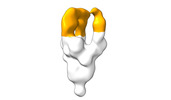
EMDB-28098: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-294

EMDB-28099: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-295

EMDB-28100: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-299

EMDB-28102: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-334
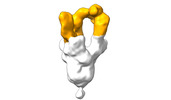
EMDB-28103: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-360

EMDB-28104: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-361

EMDB-28105: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-362

EMDB-28106: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-368

EMDB-28168: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-292

EMDB-28169: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-333
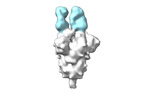
EMDB-28170: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-355
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
