-Search query
-Search result
Showing all 38 items for (author: bess & j)

EMDB-16512: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle

EMDB-16522: 
Structure of ADDoCoV-ADAH11

PDB-8c9n: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle

EMDB-29292: 
HIV-1 envelope trimer bound to one CD4 molecule on membranes

EMDB-29293: 
HIV-1 envelope trimer bound to two CD4 molecules on membranes

EMDB-29294: 
HIV-1 envelope trimer bound to three CD4 molecules on membranes

EMDB-29295: 
Consensus structure of HIV-1 envelope trimer bound to CD4 receptors on membranes

EMDB-41045: 
Unliganded HIV-1 Env on virion

EMDB-27735: 
Cryo-EM structure of SIVmac239 SOS-2P Env trimer in complex with human bNAb PGT145

PDB-8dvd: 
Cryo-EM structure of SIVmac239 SOS-2P Env trimer in complex with human bNAb PGT145

EMDB-27718: 
SIV E660.CR54 SOS-2P Env Trimer with ITS92.02

PDB-8dua: 
SIV E660.CR54 SOS-2P Env Trimer with ITS92.02

EMDB-25062: 
In-situ structure of SIV trimer

EMDB-25063: 
in situ SIVmac239-Env trimers on the surface of AT-2-inactivated virions

EMDB-25064: 
in situ SIVmac239-Env trimers on the surface of AT-2-inactivated virions

EMDB-25065: 
In-situ structure of SIVmac239-Env trimers with ITS90.03 associated

EMDB-14571: 
Negative stain 3D structure of the plastid-encoded RNA polymerase from Sinapis alba

EMDB-27631: 
SIV mac239 SOS-2P K169T Env trimer with bNAb PGT145 and jacalin

EMDB-26608: 
Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 in complex with antibody 41 - Upright conformation

EMDB-26609: 
Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 - Reversed conformation

PDB-7ums: 
Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 in complex with antibody 41 - Upright conformation

PDB-7umt: 
Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 - Reversed conformation

EMDB-12158: 
CryoEM structure of Mycobacterium tuberculosis UMP Kinase (UMPK) in complex with UDP and UTP

PDB-7bes: 
CryoEM structure of Mycobacterium tuberculosis UMP Kinase (UMPK) in complex with UDP and UTP

EMDB-32033: 
14pf microtubule decorated with EML1-GFP

EMDB-21411: 
HIV envelope glycoprotein bound with soluble CD4 (D1-D2) and antibody 17b on AT-2 treated BaL strain virus

EMDB-21412: 
Ligand-free HIV envelope glycoprotein on AT-2 treated BaL strain virus

EMDB-21413: 
HIV envelope glycoprotein bound with antibodies 10-1074 and 3BNC117 on AT-2 treated BaL strain virus
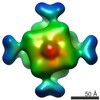
EMDB-2069: 
Octameric structure of IMPDH from Pseudomonas aeruginosa in its APO form

EMDB-2070: 
Octameric structure of IMPDH from Pseudomonas aeruginosa in presence of Mg-ATP

EMDB-1846: 
Human native spliceosomal C complex assembled on PM5 pre-mRNA.

EMDB-1847: 
Human 35S U5 snRNP

EMDB-1848: 
High salt resistant human spliceosomal C complex core

EMDB-5272: 
Molecular Structure of Unliganded Native SIVmneE11S gp120 trimer: Spike region
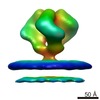
EMDB-5273: 
Molecular Structure of Unliganded Native SIVmac239 gp120 trimer: Spike region

EMDB-5274: 
Molecular Structure of Unliganded Native CP-MAC gp120 trimer: Spike region

EMDB-1246: 
Distribution and three-dimensional structure of AIDS virus envelope spikes.

EMDB-1247: 
Distribution and three-dimensional structure of AIDS virus envelope spikes.
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
