-Search query
-Search result
Showing 1 - 50 of 120 items for (author: engel & bd)
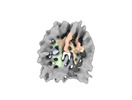
EMDB-19906: 
In situ nucleosome subtomogram average from Chlamydomonas reinhardtii
Method: subtomogram averaging / : Michael AK, Righetto RD, Obr M, van der Stappen P, Lamm L, Khavnekar S, Kotecha A, Engel BD

EMDB-13477: 
Small subunit of the Chlamydomonas reinhardtii mitoribosome
Method: single particle / : Waltz F, Soufari H, Hashem Y

EMDB-16451: 
Subtomogram average of the T. kivui 70S ribosome in situ
Method: subtomogram averaging / : Righetto RD, Dietrich HM, Kumar A, Wietrzynski W, Schuller SK, Trischler R, Wagner J, Schwarz FM, Mueller V, Schuller JM, Engel BD

EMDB-15252: 
In situ subtomogram average of the C. reinhardtii stellate at the ciliary transition zone
Method: subtomogram averaging / : van den Hoek HG, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Engel BD

EMDB-15253: 
In situ subtomogram average of the C. reinhardtii Y-link at the ciliary transition zone
Method: subtomogram averaging / : van den Hoek HG, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Engel BD

EMDB-15254: 
In situ subtomogram average of the C. reinhardtii MTD sleeve at the ciliary transition zone
Method: subtomogram averaging / : van den Hoek HG, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Engel BD

EMDB-15255: 
In situ subtomogram average of the C. reinhardtii MTD at the ciliary transition zone
Method: subtomogram averaging / : van den Hoek HG, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Engel BD

EMDB-15256: 
Composite map of the C. reinhardtii ciliary transition zone (structures attached to a single MTD) from in situ subtomogram averaging
Method: subtomogram averaging / : van den Hoek HG, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Engel BD

EMDB-15257: 
Composite map of the C. reinhardtii ciliary transition zone (full 9-fold assembly) from in situ subtomogram averaging
Method: subtomogram averaging / : van den Hoek HG, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Engel BD

EMDB-15258: 
In situ subtomogram average of C. reinhardtii IFT-B at the ciliary transition zone
Method: subtomogram averaging / : van den Hoek HG, Jordan MA, Alvarez Viar G, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Pigino G, Engel BD

EMDB-15259: 
In situ subtomogram average of C. reinhardtii IFT-A at the ciliary transition zone
Method: subtomogram averaging / : van den Hoek HG, Jordan MA, Alvarez Viar G, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Pigino G, Engel BD

EMDB-15260: 
In situ subtomogram average of C. reinhardtii dynein-1b at the ciliary transition zone
Method: subtomogram averaging / : van den Hoek HG, Jordan MA, Alvarez Viar G, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Pigino G, Engel BD

EMDB-15261: 
Composite map of the C. reinhardtii IFT (segment of a fully assembled train) at the ciliary transition zone
Method: subtomogram averaging / : van den Hoek HG, Jordan MA, Alvarez Viar G, Righetto RD, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Pigino G, Engel BD

EMDB-15262: 
Tomogram #3 of the C. reinhardtii ciliary transition zone
Method: electron tomography / : van den Hoek HG, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Engel BD

EMDB-15263: 
Tomogram #12 of the C. reinhardtii ciliary transition zone
Method: electron tomography / : van den Hoek HG, Schaffer M, Erdmann PS, Wan WN, Plitzko JM, Baumeister W, Engel BD
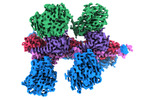
EMDB-14169: 
Cryo-EM structure of Hydrogen-dependent CO2 reductase.
Method: single particle / : Dietrich HM, Righetto RD, Kumar A, Wietrzynski W, Schuller SK, Trischler R, Wagner J, Schwarz FM, Engel BD, Mueller V, Schuller JM

EMDB-15053: 
In situ structure of HDCR filaments
Method: subtomogram averaging / : Dietrich HM, Righetto RD, Kumar A, Wietrzynski W, Schuller SK, Trischler R, Wagner J, Schwarz FM, Engel BD, Mueller V, Schuller JM

EMDB-15054: 
In situ structure of the T. kivui ribosome
Method: subtomogram averaging / : Dietrich HM, Righetto RD, Kumar A, Wietrzynski W, Schuller SK, Trischler R, Wagner J, Schwarz FM, Engel BD, Mueller V, Schuller JM

EMDB-15055: 
In situ cryo-electron tomogram of a T. kivui cell 1
Method: electron tomography / : Dietrich HM, Righetto RD, Kumar A, Wietrzynski W, Schuller SK, Trischler R, Wagner J, Schwarz FM, Engel BD, Mueller V, Schuller JM

EMDB-15056: 
In situ cryo-electron tomogram of a T. kivui cell 2
Method: electron tomography / : Dietrich HM, Righetto RD, Kumar A, Wietrzynski W, Schuller SK, Trischler R, Wagner J, Schwarz FM, Engel BD, Mueller V, Schuller JM

EMDB-13480: 
Large subunit of the Chlamydomonas reinhardtii mitoribosome
Method: single particle / : Waltz F, Soufari H, Hashem Y

EMDB-13481: 
Small subunit of the Chlamydomonas mitoribosome - head focus
Method: single particle / : Waltz F, Salinas-Giege T, Englmeier R, Meichel H, Soufari H, Kuhn L, Pfeffer S, Foerster F, Engel BD, Giege P, Drouard L, Hashem Y

EMDB-13578: 
C.reinhardtii mitochondrial ribosome (in situ)
Method: subtomogram averaging / : Waltz F, Salinas-Giege T, Englmeier R, Meichel H, Soufari H, Kuhn L, Pfeffer S, Foerster F, Engel BD, Giege P, Drouard L, Hashem Y

EMDB-12324: 
In situ cryo-electron tomogram of the Synechocystis wild-type VIPP1 strain grown in high light
Method: electron tomography / : Wietrzynski W, Klumpe S, Heinz S, Spaniol B, Schaffer M, Rast A, Nickelsen J, Schroda M, Engel BD

EMDB-12325: 
In situ cryo-electron tomogram of the Synechocystis F4E VIPP1 mutant grown in high light
Method: electron tomography / : Wietrzynski W, Klumpe S, Heinz S, Spaniol B, Schaffer M, Rast A, Nickelsen J, Schroda M, Engel BD

EMDB-12326: 
In situ cryo-electron tomogram of the Synechocystis V11E VIPP1 mutant grown in high light
Method: electron tomography / : Wietrzynski W, Klumpe S, Heinz S, Spaniol B, Schaffer M, Rast A, Nickelsen J, Schroda M, Engel BD

EMDB-12327: 
In situ cryo-electron tomogram of a cluster of VIPP1-mCherry structures inside the Chlamydomonas chloroplast
Method: electron tomography / : Wietrzynski W, Klumpe S, Heinz S, Spaniol B, Schaffer M, Rast A, Nickelsen J, Schroda M, Engel BD

EMDB-12328: 
In situ cryo-electron tomogram of a cluster of VIPP1-mCherry structures inside the Chlamydomonas chloroplast
Method: electron tomography / : Wietrzynski W, Klumpe S, Heinz S, Spaniol B, Schaffer M, Rast A, Nickelsen J, Schroda M, Engel BD

EMDB-12329: 
In situ cryo-electron tomogram of a cluster of VIPP1-mCherry structures inside the Chlamydomonas chloroplast
Method: electron tomography / : Wietrzynski W, Klumpe S, Heinz S, Spaniol B, Schaffer M, Rast A, Nickelsen J, Schroda M, Engel BD

EMDB-12330: 
In situ cryo-electron tomogram of a cluster of VIPP1-mCherry structures inside the Chlamydomonas chloroplast
Method: electron tomography / : Wietrzynski W, Klumpe S, Heinz S, Spaniol B, Schaffer M, Rast A, Nickelsen J, Schroda M, Engel BD

EMDB-12710: 
Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity
Method: single particle / : Gupta TK, Klumpe S, Gries K, Strauss M, Rudack T, Schuller JM, Schroda M, Engel BD

EMDB-12711: 
Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity
Method: single particle / : Gupta TK, Klumpe S, Gries K, Strauss M, Rudack T, Schuller JM, Schroda M, Engel BD

EMDB-12712: 
Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity
Method: single particle / : Gupta TK, Klumpe S, Gries K, Strauss M, Rudack T, Schuller JM, Schroda M, Engel BD

EMDB-12713: 
Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity
Method: single particle / : Gupta TK, Klumpe S, Gries K, Strauss M, Rudack T, Schuller JM, Schroda M, Engel BD

EMDB-12714: 
Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity
Method: single particle / : Gupta TK, Klumpe S, Gries K, Strauss M, Rudack T, Schuller JM, Schroda M, Engel BD

EMDB-12749: 
In situ cryo-electron tomogram of a pyrenoid inside a Chlamydomonas reinhardtii cell
Method: electron tomography / : Cuellar LK, Schaffer M, Strauss M, Martinez-Sanchez A, Plitzko JM, Foerster F, Engel BD

EMDB-12335: 
Structure of PSII-M
Method: single particle / : Zabret J, Bohn S, Schuller SK, Arnolds O, Chan A, Tajkhorshid E, Stoll R, Engel BD, Rudack T, Schuller JM, Nowaczyk MM

EMDB-12336: 
Structure of PSII-I (PSII with Psb27, Psb28, and Psb34)
Method: single particle / : Zabret J, Bohn S, Schuller SK, Arnolds O, Chan A, Tajkhorshid E, Stoll R, Engel BD, Rudack T, Schuller JM, Nowaczyk MM

EMDB-12337: 
Structure of PSII-I prime (PSII with Psb28, and Psb34)
Method: single particle / : Zabret J, Bohn S, Schuller SK, Arnolds O, Chan A, Tajkhorshid E, Stoll R, Engel BD, Rudack T, Schuller JM, Nowaczyk MM

EMDB-22308: 
EPYC1(49-72)-bound Rubisco
Method: single particle / : Chou H, Matthies D, He S, Jonikas MC, Yu Z

EMDB-22401: 
Rubisco in the apo state
Method: single particle / : Chou H, Matthies D, He S, Jonikas MC, Yu Z

EMDB-22462: 
EPYC1(106-135) peptide-bound Rubisco
Method: single particle / : Chou H, Matthies D, He S, Jonikas MC, Yu Z
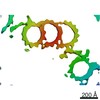
EMDB-10726: 
Electron cryo-tomography reconstruction and subvolume averaging of the A-C linker from the end of the proximal region of Paramecium tetraurelia basal body
Method: subtomogram averaging / : Guichard P, Hamel V, Le Guennec M

EMDB-10727: 
Electron cryo-tomography reconstruction and subvolume averaging of the A-C linker from the start of the proximal region of Paramecium tetraurelia basal body
Method: subtomogram averaging / : Guichard P, Hamel V, Le Guennec M

EMDB-10728: 
Electron cryo-tomography reconstruction and subvolume averaging of the microtubule triplet from the end of the proximal region of Paramecium tetraurelia basal body
Method: subtomogram averaging / : Guichard P, Hamel V, Le Guennec M

EMDB-10729: 
Electron cryo-tomography reconstruction and subvolume averaging of the microtubule triplet from the start of the proximal region of Paramecium tetraurelia basal body
Method: subtomogram averaging / : Guichard P, Hamel V, Le Guennec M

EMDB-10780: 
In situ cryo-electron tomogram of the Chlamydomonas chloroplast (Volta phase plate, bin4)
Method: electron tomography / : Schaffer M, Wietrzynski W, Tegunov D, Albert S, Plitzko J, Baumeister W, Engel BD

EMDB-10781: 
In situ cryo-electron tomogram of the Chlamydomonas chloroplast (Volta phase plate, bin4)
Method: electron tomography / : Schaffer M, Wietrzynski W, Tegunov D, Albert S, Plitzko J, Baumeister W, Engel BD

EMDB-10782: 
In situ cryo-electron tomogram of the Chlamydomonas chloroplast (Volta phase plate, bin4)
Method: electron tomography / : Schaffer M, Wietrzynski W, Tegunov D, Albert S, Plitzko J, Baumeister W, Engel BD

EMDB-10783: 
In situ cryo-electron tomogram of the Chlamydomonas chloroplast (Volta phase plate, bin4)
Method: electron tomography / : Schaffer M, Wietrzynski W, Tegunov D, Albert S, Plitzko J, Baumeister W, Engel BD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
