+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5kc2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Negative stain structure of Vps15/Vps34 complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  ENDOCYTOSIS / ENDOCYTOSIS /  autophagy / phosphatidylinositol 3-kinase (PtdIns3K) autophagy / phosphatidylinositol 3-kinase (PtdIns3K) | ||||||
| Function / homology |  Function and homology information Function and homology informationSynthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the early endosome membrane / RHO GTPases Activate NADPH Oxidases / autophagy of peroxisome / nucleus-vacuole junction / Synthesis of PIPs at the Golgi membrane / vacuole-isolation membrane contact site / vacuole inheritance / phosphatidylinositol 3-kinase complex, class III, type II / phosphatidylinositol 3-kinase complex, class III, type I ...Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the early endosome membrane / RHO GTPases Activate NADPH Oxidases / autophagy of peroxisome / nucleus-vacuole junction / Synthesis of PIPs at the Golgi membrane / vacuole-isolation membrane contact site / vacuole inheritance / phosphatidylinositol 3-kinase complex, class III, type II / phosphatidylinositol 3-kinase complex, class III, type I /  Macroautophagy / protein retention in Golgi apparatus / pexophagy / phagophore assembly site membrane / protein targeting to vacuole / late endosome to vacuole transport / fungal-type vacuole membrane / phagophore assembly site / phosphatidylinositol-mediated signaling / Macroautophagy / protein retention in Golgi apparatus / pexophagy / phagophore assembly site membrane / protein targeting to vacuole / late endosome to vacuole transport / fungal-type vacuole membrane / phagophore assembly site / phosphatidylinositol-mediated signaling /  phosphatidylinositol 3-kinase / phosphatidylinositol-3-phosphate biosynthetic process / 1-phosphatidylinositol-3-kinase activity / phosphatidylinositol phosphate biosynthetic process / phosphatidylinositol 3-kinase / phosphatidylinositol-3-phosphate biosynthetic process / 1-phosphatidylinositol-3-kinase activity / phosphatidylinositol phosphate biosynthetic process /  autophagosome assembly / autophagosome assembly /  ubiquitin binding / positive regulation of transcription elongation by RNA polymerase II / ubiquitin binding / positive regulation of transcription elongation by RNA polymerase II /  macroautophagy / macroautophagy /  autophagy / autophagy /  peroxisome / peroxisome /  endocytosis / endocytosis /  protein transport / late endosome / endosome membrane / protein transport / late endosome / endosome membrane /  non-specific serine/threonine protein kinase / non-specific serine/threonine protein kinase /  endosome / endosome /  protein kinase activity / protein kinase activity /  phosphorylation / phosphorylation /  Golgi membrane / protein serine kinase activity / protein serine/threonine kinase activity / Golgi membrane / protein serine kinase activity / protein serine/threonine kinase activity /  mitochondrion / mitochondrion /  ATP binding / ATP binding /  membrane / membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 28 Å negative staining / Resolution: 28 Å | ||||||
 Authors Authors | Kirsten, M.L. / Zhang, L. / Ohashi, Y. / Perisic, O. / Williams, R.L. / Sachse, C. | ||||||
 Citation Citation |  Journal: Autophagy / Year: 2016 Journal: Autophagy / Year: 2016Title: Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex. Authors: Yohei Ohashi / Nicolas Soler / Miguel García Ortegón / Lufei Zhang / Marie L Kirsten / Olga Perisic / Glenn R Masson / John E Burke / Arjen J Jakobi / Apostolos A Apostolakis / Christopher ...Authors: Yohei Ohashi / Nicolas Soler / Miguel García Ortegón / Lufei Zhang / Marie L Kirsten / Olga Perisic / Glenn R Masson / John E Burke / Arjen J Jakobi / Apostolos A Apostolakis / Christopher M Johnson / Maki Ohashi / Nicholas T Ktistakis / Carsten Sachse / Roger L Williams /   Abstract: The phosphatidylinositol 3-kinase Vps34 is part of several protein complexes. The structural organization of heterotetrameric complexes is starting to emerge, but little is known about organization ...The phosphatidylinositol 3-kinase Vps34 is part of several protein complexes. The structural organization of heterotetrameric complexes is starting to emerge, but little is known about organization of additional accessory subunits that interact with these assemblies. Combining hydrogen-deuterium exchange mass spectrometry (HDX-MS), X-ray crystallography and electron microscopy (EM), we have characterized Atg38 and its human ortholog NRBF2, accessory components of complex I consisting of Vps15-Vps34-Vps30/Atg6-Atg14 (yeast) and PIK3R4/VPS15-PIK3C3/VPS34-BECN1/Beclin 1-ATG14 (human). HDX-MS shows that Atg38 binds the Vps30-Atg14 subcomplex of complex I, using mainly its N-terminal MIT domain and bridges the coiled-coil I regions of Atg14 and Vps30 in the base of complex I. The Atg38 C-terminal domain is important for localization to the phagophore assembly site (PAS) and homodimerization. Our 2.2 Å resolution crystal structure of the Atg38 C-terminal homodimerization domain shows 2 segments of α-helices assembling into a mushroom-like asymmetric homodimer with a 4-helix cap and a parallel coiled-coil stalk. One Atg38 homodimer engages a single complex I. This is in sharp contrast to human NRBF2, which also forms a homodimer, but this homodimer can bridge 2 complex I assemblies. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5kc2.cif.gz 5kc2.cif.gz | 318.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5kc2.ent.gz pdb5kc2.ent.gz | 223 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5kc2.json.gz 5kc2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/kc/5kc2 https://data.pdbj.org/pub/pdb/validation_reports/kc/5kc2 ftp://data.pdbj.org/pub/pdb/validation_reports/kc/5kc2 ftp://data.pdbj.org/pub/pdb/validation_reports/kc/5kc2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 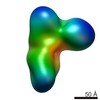 8235MC  5kc1C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 101047.180 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Gene: VPS34, END12, PEP15, VPL7, VPT29, YLR240W, L9672.10 / Production host:   Saccharomyces cerevisiae (brewer's yeast) / References: UniProt: P22543, Saccharomyces cerevisiae (brewer's yeast) / References: UniProt: P22543,  phosphatidylinositol 3-kinase phosphatidylinositol 3-kinase |
|---|---|
| #2: Protein | Mass: 166550.000 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Gene: VPS15, GRD8, VAC4, VPL19, YBR097W, YBR0825 / Production host:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)References: UniProt: P22219,  non-specific serine/threonine protein kinase non-specific serine/threonine protein kinase |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Vps15/34 / Type: COMPLEX Details: Subcomplex of Vps34 phosphatidylinositol 3-kinase (PtdIns3K) complex I (yeast) Entity ID: all / Source: RECOMBINANT | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | ||||||||||||||||||||
| Source (recombinant) | Organism:   Saccharomyces cerevisiae (brewer's yeast) / Plasmid Saccharomyces cerevisiae (brewer's yeast) / Plasmid : pYO225 : pYO225 | ||||||||||||||||||||
| Buffer solution | pH: 8.8 | ||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||
| Specimen | Conc.: 5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : YES / Vitrification applied : YES / Vitrification applied : NO : NO | ||||||||||||||||||||
| EM staining | Type: NEGATIVE / Details: droplet technique / Material: Uranyl acetate | ||||||||||||||||||||
| Specimen support |
|
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI POLARA 300 |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 120 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 120 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1500 nm / Nominal defocus min: 1000 nm Bright-field microscopy / Nominal defocus max: 1500 nm / Nominal defocus min: 1000 nm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: OTHER |
| Image recording | Average exposure time: 1 sec. / Electron dose: 20 e/Å2 / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Num. of grids imaged: 2 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING ONLY | ||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 14172 | ||||||||||||||||||||||||||||
3D reconstruction | Resolution: 28 Å / Resolution method: FSC 0.5 CUT-OFF / Num. of particles: 14172 / Algorithm: BACK PROJECTION / Symmetry type: POINT |
 Movie
Movie Controller
Controller










 PDBj
PDBj




