+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8764 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy structure of a TRPML3 ion channel | |||||||||
 Map data Map data | TRPML3 ion channel | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  transient receptor potential channel / mucolipin / transient receptor potential channel / mucolipin /  ion channel / ion channel /  membrane transport / TRPML / membrane transport / TRPML /  TRP channel / TRP channel /  calcium channel / calcium channel /  PIP2 / PI(3 / 5)P2 / lipid-gated channel / mucolipidosis / lysosomal ion channel / PIP2 / PI(3 / 5)P2 / lipid-gated channel / mucolipidosis / lysosomal ion channel /  lysosome / lysosome /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationstereocilium membrane / : / intracellularly phosphatidylinositol-3,5-bisphosphate-gated monatomic cation channel activity / inner ear auditory receptor cell differentiation / phosphatidylinositol-3,5-bisphosphate binding / monoatomic cation transmembrane transport / autophagosome membrane / locomotory behavior / late endosome membrane / early endosome membrane ...stereocilium membrane / : / intracellularly phosphatidylinositol-3,5-bisphosphate-gated monatomic cation channel activity / inner ear auditory receptor cell differentiation / phosphatidylinositol-3,5-bisphosphate binding / monoatomic cation transmembrane transport / autophagosome membrane / locomotory behavior / late endosome membrane / early endosome membrane / protein homotetramerization / membrane => GO:0016020 Similarity search - Function | |||||||||
| Biological species |   Callithrix jacchus (white-tufted-ear marmoset) Callithrix jacchus (white-tufted-ear marmoset) | |||||||||
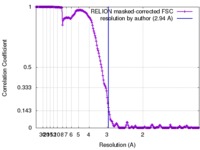
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.94 Å cryo EM / Resolution: 2.94 Å | |||||||||
 Authors Authors | Hirschi M / Herzik MA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Authors: Marscha Hirschi / Mark A Herzik / Jinhong Wie / Yang Suo / William F Borschel / Dejian Ren / Gabriel C Lander / Seok-Yong Lee /  Abstract: The modulation of ion channel activity by lipids is increasingly recognized as a fundamental component of cellular signalling. The transient receptor potential mucolipin (TRPML) channel family ...The modulation of ion channel activity by lipids is increasingly recognized as a fundamental component of cellular signalling. The transient receptor potential mucolipin (TRPML) channel family belongs to the TRP superfamily and is composed of three members: TRPML1-TRPML3. TRPMLs are the major Ca-permeable channels on late endosomes and lysosomes (LEL). They regulate the release of Ca from organelles, which is important for various physiological processes, including organelle trafficking and fusion. Loss-of-function mutations in the MCOLN1 gene, which encodes TRPML1, cause the neurodegenerative lysosomal storage disorder mucolipidosis type IV, and a gain-of-function mutation (Ala419Pro) in TRPML3 gives rise to the varitint-waddler (Va) mouse phenotype. Notably, TRPML channels are activated by the low-abundance and LEL-enriched signalling lipid phosphatidylinositol-3,5-bisphosphate (PtdIns(3,5)P), whereas other phosphoinositides such as PtdIns(4,5)P, which is enriched in plasma membranes, inhibit TRPMLs. Conserved basic residues at the N terminus of the channel are important for activation by PtdIns(3,5)P and inhibition by PtdIns(4,5)P. However, owing to a lack of structural information, the mechanism by which TRPML channels recognize PtdIns(3,5)P and increase their Ca conductance remains unclear. Here we present the cryo-electron microscopy (cryo-EM) structure of a full-length TRPML3 channel from the common marmoset (Callithrix jacchus) at an overall resolution of 2.9 Å. Our structure reveals not only the molecular basis of ion conduction but also the unique architecture of TRPMLs, wherein the voltage sensor-like domain is linked to the pore via a cytosolic domain that we term the mucolipin domain. Combined with functional studies, these data suggest that the mucolipin domain is responsible for PtdIns(3,5)P binding and subsequent channel activation, and that it acts as a 'gating pulley' for lipid-dependent TRPML gating. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8764.map.gz emd_8764.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8764-v30.xml emd-8764-v30.xml emd-8764.xml emd-8764.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8764_fsc.xml emd_8764_fsc.xml | 17.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_8764.png emd_8764.png | 224.1 KB | ||
| Filedesc metadata |  emd-8764.cif.gz emd-8764.cif.gz | 5.8 KB | ||
| Others |  emd_8764_additional.map.gz emd_8764_additional.map.gz emd_8764_half_map_1.map.gz emd_8764_half_map_1.map.gz emd_8764_half_map_2.map.gz emd_8764_half_map_2.map.gz | 59.3 MB 404.3 MB 404.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8764 http://ftp.pdbj.org/pub/emdb/structures/EMD-8764 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8764 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8764 | HTTPS FTP |
-Related structure data
| Related structure data |  5w3sMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8764.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8764.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRPML3 ion channel | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.655 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: TRPML3 ion channel
| File | emd_8764_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRPML3 ion channel | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: TRPML3 ion channel
| File | emd_8764_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRPML3 ion channel | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: TRPML3 ion channel
| File | emd_8764_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TRPML3 ion channel | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Transient Receptor Potential Mucolipin 3
| Entire | Name: Transient Receptor Potential Mucolipin 3 |
|---|---|
| Components |
|
-Supramolecule #1: Transient Receptor Potential Mucolipin 3
| Supramolecule | Name: Transient Receptor Potential Mucolipin 3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Callithrix jacchus (white-tufted-ear marmoset) Callithrix jacchus (white-tufted-ear marmoset) |
| Molecular weight | Theoretical: 260 KDa |
-Macromolecule #1: Mucolipin-3 isoform 1
| Macromolecule | Name: Mucolipin-3 isoform 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Callithrix jacchus (white-tufted-ear marmoset) Callithrix jacchus (white-tufted-ear marmoset) |
| Molecular weight | Theoretical: 65.092301 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MANPEIVISS CSSHEEENRC NFNQHTSPSE ELLLEDQMRR KLKFFFMNPC EKFWARGRKP WKLAIQILKI AMVTIQLVLF GLSNQMVVA FKEENTVAFK HLFLKGYIDR MDDTYAVYTQ SDVYDQIIFA VNQYLQLYQV SVGNHAYENK GTDQSAMAIC Q HFYKRGNI ...String: MANPEIVISS CSSHEEENRC NFNQHTSPSE ELLLEDQMRR KLKFFFMNPC EKFWARGRKP WKLAIQILKI AMVTIQLVLF GLSNQMVVA FKEENTVAFK HLFLKGYIDR MDDTYAVYTQ SDVYDQIIFA VNQYLQLYQV SVGNHAYENK GTDQSAMAIC Q HFYKRGNI YPGNDTFDID PEIETDCFFV EPDEPFHIGT PAENKLNLTL DFHRLLTVEL QFKLKAINLQ TVRHQELPDC YD FTLTITF DNKAHSGRIK ISLDNDISIR ECKDWHVSGS IQKNTHNMMI FDAFVILTCL VSLILCIRSV ISGLQLQQEF VNF FLLHYK KDVSVSDQME FVNGWYIMII ISDILTIIGS ILKMEIQAKS LTSYDVCSIL LGTSTMLVWL GVIRYLGFFA KYNL LILTL QAALPNVIRF CCCAAMIYLG YCFCGWIVLG PYHNKFRSLN MVSECLFSLI NGDDMFATFA KMQQKSYLVW LFSRI YLYS FISLFIYMIL SLFIALITDT YETIKHYQQD GFPETELRTF ISECKDLPNS GKFRLEDDPP VSLFCCCKKS NLEVLF Q UniProtKB: Mucolipin-3 |
-Macromolecule #2: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 2 / Number of copies: 12 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #3: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 3 / Number of copies: 6 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #4: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 4 / Number of copies: 4 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 12.0 sec. / Average electron dose: 63.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)