[English] 日本語
 Yorodumi
Yorodumi- EMDB-8707: Negative stain reconstruction of Endoplasmic Reticulum HSP40 co-c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8707 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
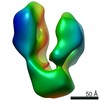
| Title | Negative stain reconstruction of Endoplasmic Reticulum HSP40 co-chaperone native ERdj3 tetramer. | |||||||||
 Map data Map data | Negative stain reconstruction of native ERdj3 tetramer | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationXBP1(S) activates chaperone genes /  misfolded protein binding / positive regulation of ATP-dependent activity / protein folding chaperone complex / IRE1-mediated unfolded protein response / protein maturation / unfolded protein binding / misfolded protein binding / positive regulation of ATP-dependent activity / protein folding chaperone complex / IRE1-mediated unfolded protein response / protein maturation / unfolded protein binding /  protein folding / protein folding /  endoplasmic reticulum lumen / endoplasmic reticulum lumen /  endoplasmic reticulum / endoplasmic reticulum /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
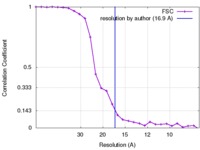
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 16.9 Å negative staining / Resolution: 16.9 Å | |||||||||
 Authors Authors | Chowdhury S / Lander GC / Chen K-C / Qu S / Noxon IC / Schonhoft JD / Plate L / Powers ET / Kelly JW / Wiseman RL | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Structures of Qβ virions, virus-like particles, and the Qβ-MurA complex reveal internal coat proteins and the mechanism of host lysis. Authors: Zhicheng Cui / Karl V Gorzelnik / Jeng-Yih Chang / Carrie Langlais / Joanita Jakana / Ry Young / Junjie Zhang /  Abstract: In single-stranded RNA bacteriophages (ssRNA phages) a single copy of the maturation protein binds the genomic RNA (gRNA) and is required for attachment of the phage to the host pilus. For the ...In single-stranded RNA bacteriophages (ssRNA phages) a single copy of the maturation protein binds the genomic RNA (gRNA) and is required for attachment of the phage to the host pilus. For the canonical Qβ the maturation protein, A, has an additional role as the lysis protein, by its ability to bind and inhibit MurA, which is involved in peptidoglycan biosynthesis. Here, we determined structures of Qβ virions, virus-like particles, and the Qβ-MurA complex using single-particle cryoelectron microscopy, at 4.7-Å, 3.3-Å, and 6.1-Å resolutions, respectively. We identified the outer surface of the β-region in A as the MurA-binding interface. Moreover, the pattern of MurA mutations that block Qβ lysis and the conformational changes of MurA that facilitate A binding were found to be due to the intimate fit between A and the region encompassing the closed catalytic cleft of substrate-liganded MurA. Additionally, by comparing the Qβ virion with Qβ virus-like particles that lack a maturation protein, we observed a structural rearrangement in the capsid coat proteins that is required to package the viral gRNA in its dominant conformation. Unexpectedly, we found a coat protein dimer sequestered in the interior of the virion. This coat protein dimer binds to the gRNA and interacts with the buried α-region of A, suggesting that it is sequestered during the early stage of capsid formation to promote the gRNA condensation required for genome packaging. These internalized coat proteins are the most asymmetrically arranged major capsid proteins yet observed in virus structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8707.map.gz emd_8707.map.gz | 783.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8707-v30.xml emd-8707-v30.xml emd-8707.xml emd-8707.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8707_fsc.xml emd_8707_fsc.xml | 2.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_8707.png emd_8707.png | 39.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8707 http://ftp.pdbj.org/pub/emdb/structures/EMD-8707 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8707 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8707 | HTTPS FTP |
-Related structure data
| Related structure data |  8708C  8709C  8710C  8711C  5vlyC  5vlzC  5vm7C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8707.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8707.map.gz / Format: CCP4 / Size: 844.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain reconstruction of native ERdj3 tetramer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tetrameric complex of native ERdj3 protein complex
| Entire | Name: Tetrameric complex of native ERdj3 protein complex |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric complex of native ERdj3 protein complex
| Supramolecule | Name: Tetrameric complex of native ERdj3 protein complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant cell: SHuffle T7 Express Escherichia coli (E. coli) / Recombinant cell: SHuffle T7 Express |
| Molecular weight | Theoretical: 153 KDa |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.025 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 50 mM HEPES, 300 mM NaCl, 1% (v/v)Glycerol |
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Sample absorbed on carbon surface was stained with 2% (w/v) Uranyl Formate solution, and finally air-dried after blotting off excess stain. |
| Grid | Model: 400 mesh Cu-Rh maxtaform grids (Electron Microscopy Sciences) that were coated with a thin layer of amorphous carbon. Material: COPPER/RHODIUM / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: 15mA current |
| Details | Mono dispersed protein solution |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 Bright-field microscopy / Cs: 2.2 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Temperature | Min: 293.0 K / Max: 295.0 K |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 15.6 µm / Number grids imaged: 2 / Number real images: 980 / Average exposure time: 0.79 sec. / Average electron dose: 35.0 e/Å2 Details: Automated image acquisition software Leginon was used for data collection. |
 Movie
Movie Controller
Controller