+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8699 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EBOV GPdMuc in complex with ADI-15742 Fab | |||||||||
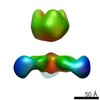
 Map data Map data | Recombinant Ebola virus glycoprotein in complex with recombinant human IgG1 Fab ADI-15742 from survivor in infection. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 20.8 Å negative staining / Resolution: 20.8 Å | |||||||||
 Authors Authors | Murin CD / Ward AB / Turner HL | |||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: Antibodies from a Human Survivor Define Sites of Vulnerability for Broad Protection against Ebolaviruses. Authors: Anna Z Wec / Andrew S Herbert / Charles D Murin / Elisabeth K Nyakatura / Dafna M Abelson / J Maximilian Fels / Shihua He / Rebekah M James / Marc-Antoine de La Vega / Wenjun Zhu / Russell R ...Authors: Anna Z Wec / Andrew S Herbert / Charles D Murin / Elisabeth K Nyakatura / Dafna M Abelson / J Maximilian Fels / Shihua He / Rebekah M James / Marc-Antoine de La Vega / Wenjun Zhu / Russell R Bakken / Eileen Goodwin / Hannah L Turner / Rohit K Jangra / Larry Zeitlin / Xiangguo Qiu / Jonathan R Lai / Laura M Walker / Andrew B Ward / John M Dye / Kartik Chandran / Zachary A Bornholdt /   Abstract: Experimental monoclonal antibody (mAb) therapies have shown promise for treatment of lethal Ebola virus (EBOV) infections, but their species-specific recognition of the viral glycoprotein (GP) has ...Experimental monoclonal antibody (mAb) therapies have shown promise for treatment of lethal Ebola virus (EBOV) infections, but their species-specific recognition of the viral glycoprotein (GP) has limited their use against other divergent ebolaviruses associated with human disease. Here, we mined the human immune response to natural EBOV infection and identified mAbs with exceptionally potent pan-ebolavirus neutralizing activity and protective efficacy against three virulent ebolaviruses. These mAbs recognize an inter-protomer epitope in the GP fusion loop, a critical and conserved element of the viral membrane fusion machinery, and neutralize viral entry by targeting a proteolytically primed, fusion-competent GP intermediate (GP) generated in host cell endosomes. Only a few somatic hypermutations are required for broad antiviral activity, and germline-approximating variants display enhanced GP recognition, suggesting that such antibodies could be elicited more efficiently with suitably optimized GP immunogens. Our findings inform the development of both broadly effective immunotherapeutics and vaccines against filoviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8699.map.gz emd_8699.map.gz | 720.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8699-v30.xml emd-8699-v30.xml emd-8699.xml emd-8699.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8699.png emd_8699.png | 38.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8699 http://ftp.pdbj.org/pub/emdb/structures/EMD-8699 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8699 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8699 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8699.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8699.map.gz / Format: CCP4 / Size: 1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Recombinant Ebola virus glycoprotein in complex with recombinant human IgG1 Fab ADI-15742 from survivor in infection. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : EBOV GPdMuc:ADI-15742 Fab
| Entire | Name: EBOV GPdMuc:ADI-15742 Fab |
|---|---|
| Components |
|
-Supramolecule #1: EBOV GPdMuc:ADI-15742 Fab
| Supramolecule | Name: EBOV GPdMuc:ADI-15742 Fab / type: complex / ID: 1 / Parent: 0 Details: Complex of recombinant mucin-deleted Ebola virus glycoprotein and recombinant human survivor antibody IgG1 Fab ADI-15742. |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: 293F / Recombinant plasmid: pPPI4 Homo sapiens (human) / Recombinant cell: 293F / Recombinant plasmid: pPPI4 |
-Supramolecule #2: EBOV GPdMuc
| Supramolecule | Name: EBOV GPdMuc / type: complex / ID: 2 / Parent: 1 Details: Mucin-delected Ebola virus glycoprotein recombinantly expressed in mammalian cells, 293F |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: 293F / Recombinant plasmid: pPPI4 Homo sapiens (human) / Recombinant cell: 293F / Recombinant plasmid: pPPI4 |
-Supramolecule #3: ADI-15742 Fab
| Supramolecule | Name: ADI-15742 Fab / type: complex / ID: 3 / Parent: 1 Details: Fab fragment generated by proteolytic cleavage of IgG antibody |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Nicotiana benthamiana (plant) Nicotiana benthamiana (plant) |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.03 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: Solutions were made from a 10X stock and filtered sterilized. | |||||||||
| Staining | Type: NEGATIVE / Material: Uranyl Formate Details: Negatively stained EM specimens were prepared using a side-blotting technique and uranyl-formate stain. | |||||||||
| Grid | Model: Protochips / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER | |||||||||
| Details | Monodisperse sample prepared by negative stain. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.0 µm Bright-field microscopy / Nominal defocus max: 1.0 µm |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
| Image recording | Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Number grids imaged: 1 / Number real images: 74 / Average electron dose: 2.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 12705 |
|---|---|
| Startup model | Type of model: OTHER / Details: common lines model |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: EMAN (ver. 2.2) |
| Final 3D classification | Number classes: 84 / Avg.num./class: 50 / Software - Name: IMAGIC / Details: Iterative MSA/MRA. |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: EMAN (ver. 2.2) |
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 20.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN (ver. 2.2) / Number images used: 6980 ) / Resolution.type: BY AUTHOR / Resolution: 20.8 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN (ver. 2.2) / Number images used: 6980 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















