[English] 日本語
 Yorodumi
Yorodumi- EMDB-8635: VQIINK, Structure of the amyloid-spine from microtubule associate... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8635 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | VQIINK, Structure of the amyloid-spine from microtubule associated protein tau Repeat 2 | ||||||||||||
 Map data Map data | amyloid-spine from microtubule associated protein tau Repeat 2 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  Amyloid / tau / Amyloid / tau /  Alzheimer's Disease / Alzheimer's Disease /  tauopathy / tauopathy /  MAPT / MAPT /  STRUCTURAL PROTEIN STRUCTURAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationplus-end-directed organelle transport along microtubule /  axonal transport / histone-dependent DNA binding / axonal transport / histone-dependent DNA binding /  neurofibrillary tangle assembly / positive regulation of diacylglycerol kinase activity / negative regulation of establishment of protein localization to mitochondrion / neurofibrillary tangle assembly / positive regulation of diacylglycerol kinase activity / negative regulation of establishment of protein localization to mitochondrion /  neurofibrillary tangle / positive regulation of protein localization to synapse / microtubule lateral binding / neurofibrillary tangle / positive regulation of protein localization to synapse / microtubule lateral binding /  tubulin complex ...plus-end-directed organelle transport along microtubule / tubulin complex ...plus-end-directed organelle transport along microtubule /  axonal transport / histone-dependent DNA binding / axonal transport / histone-dependent DNA binding /  neurofibrillary tangle assembly / positive regulation of diacylglycerol kinase activity / negative regulation of establishment of protein localization to mitochondrion / neurofibrillary tangle assembly / positive regulation of diacylglycerol kinase activity / negative regulation of establishment of protein localization to mitochondrion /  neurofibrillary tangle / positive regulation of protein localization to synapse / microtubule lateral binding / neurofibrillary tangle / positive regulation of protein localization to synapse / microtubule lateral binding /  tubulin complex / tubulin complex /  phosphatidylinositol bisphosphate binding / main axon / regulation of long-term synaptic depression / negative regulation of kinase activity / negative regulation of tubulin deacetylation / generation of neurons / regulation of chromosome organization / positive regulation of protein localization / rRNA metabolic process / internal protein amino acid acetylation / phosphatidylinositol bisphosphate binding / main axon / regulation of long-term synaptic depression / negative regulation of kinase activity / negative regulation of tubulin deacetylation / generation of neurons / regulation of chromosome organization / positive regulation of protein localization / rRNA metabolic process / internal protein amino acid acetylation /  regulation of mitochondrial fission / regulation of mitochondrial fission /  lipoprotein particle binding / intracellular distribution of mitochondria / axonal transport of mitochondrion / axon development / lipoprotein particle binding / intracellular distribution of mitochondria / axonal transport of mitochondrion / axon development /  central nervous system neuron development / central nervous system neuron development /  regulation of microtubule polymerization / regulation of microtubule polymerization /  microtubule polymerization / minor groove of adenine-thymine-rich DNA binding / negative regulation of mitochondrial membrane potential / microtubule polymerization / minor groove of adenine-thymine-rich DNA binding / negative regulation of mitochondrial membrane potential /  dynactin binding / glial cell projection / dynactin binding / glial cell projection /  apolipoprotein binding / protein polymerization / negative regulation of mitochondrial fission / apolipoprotein binding / protein polymerization / negative regulation of mitochondrial fission /  axolemma / Caspase-mediated cleavage of cytoskeletal proteins / regulation of microtubule polymerization or depolymerization / positive regulation of axon extension / supramolecular fiber organization / Activation of AMPK downstream of NMDARs / cytoplasmic microtubule organization / regulation of microtubule cytoskeleton organization / axolemma / Caspase-mediated cleavage of cytoskeletal proteins / regulation of microtubule polymerization or depolymerization / positive regulation of axon extension / supramolecular fiber organization / Activation of AMPK downstream of NMDARs / cytoplasmic microtubule organization / regulation of microtubule cytoskeleton organization /  stress granule assembly / regulation of cellular response to heat / regulation of calcium-mediated signaling / axon cytoplasm / positive regulation of microtubule polymerization / cellular response to brain-derived neurotrophic factor stimulus / somatodendritic compartment / stress granule assembly / regulation of cellular response to heat / regulation of calcium-mediated signaling / axon cytoplasm / positive regulation of microtubule polymerization / cellular response to brain-derived neurotrophic factor stimulus / somatodendritic compartment /  synapse assembly / synapse assembly /  phosphatidylinositol binding / nuclear periphery / cellular response to nerve growth factor stimulus / positive regulation of superoxide anion generation / protein phosphatase 2A binding / phosphatidylinositol binding / nuclear periphery / cellular response to nerve growth factor stimulus / positive regulation of superoxide anion generation / protein phosphatase 2A binding /  regulation of autophagy / astrocyte activation / synapse organization / microglial cell activation / response to lead ion / regulation of autophagy / astrocyte activation / synapse organization / microglial cell activation / response to lead ion /  Hsp90 protein binding / Hsp90 protein binding /  regulation of synaptic plasticity / PKR-mediated signaling / protein homooligomerization / cytoplasmic ribonucleoprotein granule / regulation of synaptic plasticity / PKR-mediated signaling / protein homooligomerization / cytoplasmic ribonucleoprotein granule /  memory / microtubule cytoskeleton organization / memory / microtubule cytoskeleton organization /  SH3 domain binding / cellular response to reactive oxygen species / neuron projection development / activation of cysteine-type endopeptidase activity involved in apoptotic process / microtubule cytoskeleton / protein-macromolecule adaptor activity / SH3 domain binding / cellular response to reactive oxygen species / neuron projection development / activation of cysteine-type endopeptidase activity involved in apoptotic process / microtubule cytoskeleton / protein-macromolecule adaptor activity /  single-stranded DNA binding / cell-cell signaling / cellular response to heat / single-stranded DNA binding / cell-cell signaling / cellular response to heat /  cell body / cell body /  actin binding / actin binding /  growth cone / protein-folding chaperone binding / growth cone / protein-folding chaperone binding /  double-stranded DNA binding / double-stranded DNA binding /  microtubule binding / microtubule binding /  microtubule / amyloid fibril formation / sequence-specific DNA binding / microtubule / amyloid fibril formation / sequence-specific DNA binding /  dendritic spine / learning or memory / neuron projection / nuclear speck / dendritic spine / learning or memory / neuron projection / nuclear speck /  membrane raft / membrane raft /  axon / negative regulation of gene expression / axon / negative regulation of gene expression /  dendrite / neuronal cell body / DNA damage response / dendrite / neuronal cell body / DNA damage response /  protein kinase binding / protein kinase binding /  enzyme binding / enzyme binding /  mitochondrion / mitochondrion /  DNA binding DNA bindingSimilarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method |  electron crystallography / electron crystallography /  cryo EM cryo EM | ||||||||||||
 Authors Authors | Seidler PM / Sawaya MR | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Chem / Year: 2018 Journal: Nat Chem / Year: 2018Title: Structure-based inhibitors of tau aggregation. Authors: P M Seidler / D R Boyer / J A Rodriguez / M R Sawaya / D Cascio / K Murray / T Gonen / D S Eisenberg /  Abstract: Aggregated tau protein is associated with over 20 neurological disorders, which include Alzheimer's disease. Previous work has shown that tau's sequence segments VQIINK and VQIVYK drive its ...Aggregated tau protein is associated with over 20 neurological disorders, which include Alzheimer's disease. Previous work has shown that tau's sequence segments VQIINK and VQIVYK drive its aggregation, but inhibitors based on the structure of the VQIVYK segment only partially inhibit full-length tau aggregation and are ineffective at inhibiting seeding by full-length fibrils. Here we show that the VQIINK segment is the more powerful driver of tau aggregation. Two structures of this segment determined by the cryo-electron microscopy method micro-electron diffraction explain its dominant influence on tau aggregation. Of practical significance, the structures lead to the design of inhibitors that not only inhibit tau aggregation but also inhibit the ability of exogenous full-length tau fibrils to seed intracellular tau in HEK293 biosensor cells into amyloid. We also raise the possibility that the two VQIINK structures represent amyloid polymorphs of tau that may account for a subset of prion-like strains of tau. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8635.map.gz emd_8635.map.gz | 225.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8635-v30.xml emd-8635-v30.xml emd-8635.xml emd-8635.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8635.png emd_8635.png | 101 KB | ||
| Filedesc metadata |  emd-8635.cif.gz emd-8635.cif.gz | 4.4 KB | ||
| Filedesc structureFactors |  emd_8635_sf.cif.gz emd_8635_sf.cif.gz | 36 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8635 http://ftp.pdbj.org/pub/emdb/structures/EMD-8635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8635 | HTTPS FTP |
-Related structure data
| Related structure data |  5v5cMC  8634C  5v5bC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8635.map.gz / Format: CCP4 / Size: 244.1 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8635.map.gz / Format: CCP4 / Size: 244.1 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | amyloid-spine from microtubule associated protein tau Repeat 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.424 Å / Y: 0.4 Å / Z: 0.402 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
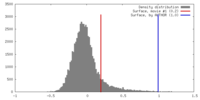
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 18 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : VQIINK Tau peptide
| Entire | Name: VQIINK Tau peptide |
|---|---|
| Components |
|
-Supramolecule #1: VQIINK Tau peptide
| Supramolecule | Name: VQIINK Tau peptide / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Microtubule-associated protein tau
| Macromolecule | Name: Microtubule-associated protein tau / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 714.873 Da |
| Sequence | String: VQIINK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  electron crystallography electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | 3D micro-crystal |
| Crystal formation | Temperature: 291.0 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Camera length: 730 mm / Camera length: 730 mm |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 0.1 e/Å2 |
- Image processing
Image processing
| Crystallography statistics | Number intensities measured: 5454 / Number structure factors: 1226 / Fourier space coverage: 86.8 / R sym: 23.9 / R merge: 23.9 / Overall phase error: 0.1 / Overall phase residual: 0.1 / Phase error rejection criteria: 0.1 / High resolution: 1.25 Å Details: This is a crystallography experiment. Phases were not measured. Shell - Shell ID: 1 / Shell - High resolution: 1.25 Å / Shell - Low resolution: 1.31 Å / Shell - Number structure factors: 106 / Shell - Phase residual: 0.1 / Shell - Fourier space coverage: 71.6 / Shell - Multiplicity: 2.5 |
|---|---|
| Molecular replacement | Software - Name: Phaser |
| Symmetry determination software list | Software - Name: XDS |
| Final reconstruction | Resolution method: DIFFRACTION PATTERN/LAYERLINES |
| Merging software list | Software - Name: XSCALE |
 Movie
Movie Controller
Controller









 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)