[English] 日本語
 Yorodumi
Yorodumi- EMDB-8624: Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8624 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | ||||||||||||
 Map data Map data | Cryo EM map of the Csy-Acr complex. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | CRISPR RNA-recognition-motif (RRM) / pseudo-helical / Type 1-F CRISPR /  IMMUNE SYSTEM-RNA complex IMMUNE SYSTEM-RNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host CRISPR-cas system / maintenance of CRISPR repeat elements / defense response to virus /  endonuclease activity / endonuclease activity /  Hydrolases; Acting on ester bonds / Hydrolases; Acting on ester bonds /  RNA binding RNA bindingSimilarity search - Function | ||||||||||||
| Biological species |   Pseudomonas aeruginosa UCBPP-PA14 (bacteria) / Pseudomonas aeruginosa UCBPP-PA14 (bacteria) /   Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) / Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) /  Pseudomonas phage JBD30 (virus) / Pseudomonas phage JBD30 (virus) /  Pseudomonas phage D3112 (virus) Pseudomonas phage D3112 (virus) | ||||||||||||
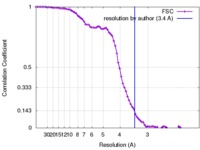
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Chowdhury S / Carter J | ||||||||||||
| Funding support |  United States, United States,  Canada, 3 items Canada, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex. Authors: Saikat Chowdhury / Joshua Carter / MaryClare F Rollins / Sarah M Golden / Ryan N Jackson / Connor Hoffmann / Lyn'Al Nosaka / Joseph Bondy-Denomy / Karen L Maxwell / Alan R Davidson / ...Authors: Saikat Chowdhury / Joshua Carter / MaryClare F Rollins / Sarah M Golden / Ryan N Jackson / Connor Hoffmann / Lyn'Al Nosaka / Joseph Bondy-Denomy / Karen L Maxwell / Alan R Davidson / Elizabeth R Fischer / Gabriel C Lander / Blake Wiedenheft /   Abstract: Genetic conflict between viruses and their hosts drives evolution and genetic innovation. Prokaryotes evolved CRISPR-mediated adaptive immune systems for protection from viral infection, and viruses ...Genetic conflict between viruses and their hosts drives evolution and genetic innovation. Prokaryotes evolved CRISPR-mediated adaptive immune systems for protection from viral infection, and viruses have evolved diverse anti-CRISPR (Acr) proteins that subvert these immune systems. The adaptive immune system in Pseudomonas aeruginosa (type I-F) relies on a 350 kDa CRISPR RNA (crRNA)-guided surveillance complex (Csy complex) to bind foreign DNA and recruit a trans-acting nuclease for target degradation. Here, we report the cryo-electron microscopy (cryo-EM) structure of the Csy complex bound to two different Acr proteins, AcrF1 and AcrF2, at an average resolution of 3.4 Å. The structure explains the molecular mechanism for immune system suppression, and structure-guided mutations show that the Acr proteins bind to residues essential for crRNA-mediated detection of DNA. Collectively, these data provide a snapshot of an ongoing molecular arms race between viral suppressors and the immune system they target. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8624.map.gz emd_8624.map.gz | 5.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8624-v30.xml emd-8624-v30.xml emd-8624.xml emd-8624.xml | 33.9 KB 33.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8624_fsc.xml emd_8624_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_8624.png emd_8624.png | 175.4 KB | ||
| Filedesc metadata |  emd-8624.cif.gz emd-8624.cif.gz | 9 KB | ||
| Others |  emd_8624_additional_1.map.gz emd_8624_additional_1.map.gz emd_8624_additional_2.map.gz emd_8624_additional_2.map.gz emd_8624_half_map_1.map.gz emd_8624_half_map_1.map.gz emd_8624_half_map_2.map.gz emd_8624_half_map_2.map.gz | 1.9 MB 31 MB 4.8 MB 4.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8624 http://ftp.pdbj.org/pub/emdb/structures/EMD-8624 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8624 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8624 | HTTPS FTP |
-Related structure data
| Related structure data |  5uz9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8624.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8624.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo EM map of the Csy-Acr complex. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Focused map of the "tail" region of Csy-Acr...
| File | emd_8624_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map of the "tail" region of Csy-Acr complex, comprising of Cas5, Cas8 and Acr2 subunits. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened Cryo EM map of the Csy-Acr complex.
| File | emd_8624_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened Cryo EM map of the Csy-Acr complex. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2 of the Csy-Acr complex.
| File | emd_8624_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 of the Csy-Acr complex. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1 of the Csy-Acr complex.
| File | emd_8624_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 of the Csy-Acr complex. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Anti-CRISPRs AcrF1 and AcrF2 bound P. aeruginosa crRNA-guided CRI...
| Entire | Name: Anti-CRISPRs AcrF1 and AcrF2 bound P. aeruginosa crRNA-guided CRISPR surveillance(Csy)complex. |
|---|---|
| Components |
|
-Supramolecule #1: Anti-CRISPRs AcrF1 and AcrF2 bound P. aeruginosa crRNA-guided CRI...
| Supramolecule | Name: Anti-CRISPRs AcrF1 and AcrF2 bound P. aeruginosa crRNA-guided CRISPR surveillance(Csy)complex. type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Cas5F, Cas6F, Cas7F, Cas8F and CRISPR RNA (crRNA) forms the Csy surveillance complex. Cas5F, Cas8F and 5'crRNA handle forms the "tail" of the complex, and 3' crRNA stem-loop along with Cas6F ...Details: Cas5F, Cas6F, Cas7F, Cas8F and CRISPR RNA (crRNA) forms the Csy surveillance complex. Cas5F, Cas8F and 5'crRNA handle forms the "tail" of the complex, and 3' crRNA stem-loop along with Cas6F forms the "head". Six copies Cas7F along with crRNA spacer forms the "core" of the complex. |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa UCBPP-PA14 (bacteria) Pseudomonas aeruginosa UCBPP-PA14 (bacteria) |
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: CRISPR-associated protein Csy1
| Macromolecule | Name: CRISPR-associated protein Csy1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 49.194168 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLQPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EALQALSDNA EQAREWRQAF I GITTVKGA ...String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLQPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EALQALSDNA EQAREWRQAF I GITTVKGA PASHSLAKQL YFPLPGSGYH LLAPLFPTSL VHHVHALLRE ARFGDAAKAA REARSRQESW PHGFSEYPNL AI QKFGGTK PQNISQLNNE RRGENWLLPS LPPNWQRQNV NAPMRHSSVF EHDFGRTPEV SRLTRTLQRF LAKTVHNNLA IRQ RRAQLV AQICDEALQY AARLRELEPG WSATPGCQLH DAEQLWLDPL RAQTDETFLQ RRLRGDWPAE VGNRFANWLN RAVS SDSQI LGSPEAAQWS QELSKELTMF KEILEDERD UniProtKB: CRISPR-associated protein Csy1 |
-Macromolecule #2: CRISPR-associated protein Csy2
| Macromolecule | Name: CRISPR-associated protein Csy2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 36.244074 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ ...String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ RRKNQRRLTR RLLPGFALVS REALLQQHLE TLRTTLPEAT TLDALLDLCR INFEPPATSS EEEASPPDAA WQ VRDKPGW LVPIPAGYNA LSPLYLPGEV RNARDRETPL RFVENLFGLG EWLSPHRVAA LSDLLWYHHA EPDKGLYRWS TPR FVEHAI A UniProtKB: CRISPR-associated protein Csy2 |
-Macromolecule #3: CRISPR-associated protein Csy3
| Macromolecule | Name: CRISPR-associated protein Csy3 / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 37.448078 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: SKPILSTASV LAFERKLDPS DALMSAGAWA QRDASQEWPA VTVREKSVRG TISNRLKTKD RDPAKLDASI QSPNLQTVDV ANLPSDADT LKVRFTLRVL GGAGTPSACN DAAYRDKLLQ TVATYVNDQG FAELARRYAH NLANARFLWR NRVGAEAVEV R INHIRQGE ...String: SKPILSTASV LAFERKLDPS DALMSAGAWA QRDASQEWPA VTVREKSVRG TISNRLKTKD RDPAKLDASI QSPNLQTVDV ANLPSDADT LKVRFTLRVL GGAGTPSACN DAAYRDKLLQ TVATYVNDQG FAELARRYAH NLANARFLWR NRVGAEAVEV R INHIRQGE VARAWRFDAL AIGLRDFKAD AELDALAELI ASGLSGSGHV LLEVVAFARI GDGQEVFPSQ ELILDKGDKK GQ KSKTLYS VRDAAAIHSQ KIGNALRTID TWYPDEDGLG PIAVEPYGSV TSQGKAYRQP KQKLDFYTLL DNWVLRDEAP AVE QQHYVI ANLIRGGVFG EAEEK UniProtKB: CRISPR-associated protein Csy3 |
-Macromolecule #4: Anti-CRISPR protein Acr30-35
| Macromolecule | Name: Anti-CRISPR protein Acr30-35 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage JBD30 (virus) Pseudomonas phage JBD30 (virus) |
| Molecular weight | Theoretical: 8.693734 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: KFIKYLSTAH LNYMNIAVYE NGSKIKARVE NVVNGKSVGA RDFDSTEQLE SWFYGLPGSG LGRIENAMNE ISRRENP UniProtKB: Uncharacterized protein |
-Macromolecule #5: Anti-CRISPR protein 30
| Macromolecule | Name: Anti-CRISPR protein 30 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage D3112 (virus) Pseudomonas phage D3112 (virus) |
| Molecular weight | Theoretical: 10.760481 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MHHHHHHIAQ QHKDTVAACE AAEAIAIAKD QVWDGEGYTK YTFDDNSVLI QSGTTQYAMD ADDADSIKGY ADWLDDEARS AEASEIERL LESVEEE UniProtKB:  Anti-CRISPR protein 30 Anti-CRISPR protein 30 |
-Macromolecule #6: CRISPR-associated endonuclease Cas6/Csy4
| Macromolecule | Name: CRISPR-associated endonuclease Cas6/Csy4 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number:  Hydrolases; Acting on ester bonds Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 21.691848 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: FTMDHYLDIR LRPDPEFPPA QLMCVLFGKL HQALVAQGGD RIGVSFPDLD ESRSRLGERL RIHASADDLR ALLARPWLEG LRDHLQFGE PAVVPHPTPY RQVSRVQAKS NPERLRRRLM RRHDLSEEEA RKRIPDTVAR ALDLPFVTLR SQSTGQHFRL F IRHGPLQV TAEEGGFTCY GLSKGGFVPW F UniProtKB: CRISPR-associated endonuclease Cas6/Csy4 |
-Macromolecule #7: CRISPR RNA (60-MER)
| Macromolecule | Name: CRISPR RNA (60-MER) / type: rna / ID: 7 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:   Pseudomonas aeruginosa UCBPP-PA14 (bacteria) Pseudomonas aeruginosa UCBPP-PA14 (bacteria) |
| Molecular weight | Theoretical: 19.249404 KDa |
| Sequence | String: CUAAGAAAUU CACGGCGGGC UUGAUGUCCG CGUCUACCUG GUUCACUGCC GUAUAGGCAG GENBANK: GENBANK: CP000438.1 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Buffer was filtered before use. | ||||||||||||
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 5 sec. / Pretreatment - Atmosphere: OTHER Details: Holey grid was coated with an amorphous carbon film. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 98 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER Details: Sample was applied to poly-L-lysine hydrobromide pre-treated amorphous carbon film coated over a holey grid. Excess sample was blotted with Whatman-1 filter paper and plunge frozen. Manual ...Details: Sample was applied to poly-L-lysine hydrobromide pre-treated amorphous carbon film coated over a holey grid. Excess sample was blotted with Whatman-1 filter paper and plunge frozen. Manual plunge freezing was performed in a cold room.. | ||||||||||||
| Details | Sample was free from any aggregation and monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 29000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 77.0 K / Max: 79.0 K |
| Details | Objective astigmatism was corrected at nominal magnification of 29000X, using Thon rings visualized with a K2 camera. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-30 / Number grids imaged: 2 / Number real images: 2261 / Average exposure time: 6.0 sec. / Average electron dose: 46.0 e/Å2 Details: Images were collected in movie mode using Leginon automated data acquisition software. Movie frames were aligned using MotionCorr program. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Details | Top scoring five models out of two hundred models generated using Rosetta were refined using Phenix and deposited as an ensemble atomic model for the final EM map. |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-5uz9: |
-Atomic model buiding 2
| Details | The docked Cas6F structure in the "head" region of the complex was reduced to C-alpha backbone. |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-5uz9: |
-Atomic model buiding 3
| Details | Initially two copies were fitted using Chimera and then backbones and side chains were fixed using Coot, followed by real space refinement in Phenix. |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-5uz9: |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X