[English] 日本語
 Yorodumi
Yorodumi- EMDB-6794: Cryo-EM structure of zika virus complexed with Fab of a human mon... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6794 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of zika virus complexed with Fab of a human monoclonal antibody named ZKA190 at 4 degrees celsius | |||||||||
 Map data Map data | zika virus complexed with Fab of a human monoclonal antibody named ZKA190 at 4 celsius degree | |||||||||
 Sample Sample |
| |||||||||
| Biological species |    Zika virus / Zika virus /   Homo sapiens (human) Homo sapiens (human) | |||||||||
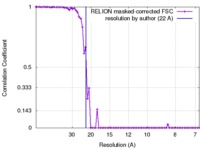
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 22.0 Å cryo EM / Resolution: 22.0 Å | |||||||||
 Authors Authors | Wang JQ / Ng TS / Fibriansah G / Lok SM | |||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: A Human Bi-specific Antibody against Zika Virus with High Therapeutic Potential. Authors: Jiaqi Wang / Marco Bardelli / Diego A Espinosa / Mattia Pedotti / Thiam-Seng Ng / Siro Bianchi / Luca Simonelli / Elisa X Y Lim / Mathilde Foglierini / Fabrizia Zatta / Stefano Jaconi / ...Authors: Jiaqi Wang / Marco Bardelli / Diego A Espinosa / Mattia Pedotti / Thiam-Seng Ng / Siro Bianchi / Luca Simonelli / Elisa X Y Lim / Mathilde Foglierini / Fabrizia Zatta / Stefano Jaconi / Martina Beltramello / Elisabetta Cameroni / Guntur Fibriansah / Jian Shi / Taylor Barca / Isabel Pagani / Alicia Rubio / Vania Broccoli / Elisa Vicenzi / Victoria Graham / Steven Pullan / Stuart Dowall / Roger Hewson / Simon Jurt / Oliver Zerbe / Karin Stettler / Antonio Lanzavecchia / Federica Sallusto / Andrea Cavalli / Eva Harris / Shee-Mei Lok / Luca Varani / Davide Corti /      Abstract: Zika virus (ZIKV), a mosquito-borne flavivirus, causes devastating congenital birth defects. We isolated a human monoclonal antibody (mAb), ZKA190, that potently cross-neutralizes multi-lineage ZIKV ...Zika virus (ZIKV), a mosquito-borne flavivirus, causes devastating congenital birth defects. We isolated a human monoclonal antibody (mAb), ZKA190, that potently cross-neutralizes multi-lineage ZIKV strains. ZKA190 is highly effective in vivo in preventing morbidity and mortality of ZIKV-infected mice. NMR and cryo-electron microscopy show its binding to an exposed epitope on DIII of the E protein. ZKA190 Fab binds all 180 E protein copies, altering the virus quaternary arrangement and surface curvature. However, ZIKV escape mutants emerged in vitro and in vivo in the presence of ZKA190, as well as of other neutralizing mAbs. To counter this problem, we developed a bispecific antibody (FIT-1) comprising ZKA190 and a second mAb specific for DII of E protein. In addition to retaining high in vitro and in vivo potencies, FIT-1 robustly prevented viral escape, warranting its development as a ZIKV immunotherapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6794.map.gz emd_6794.map.gz | 27.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6794-v30.xml emd-6794-v30.xml emd-6794.xml emd-6794.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6794_fsc.xml emd_6794_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_6794.png emd_6794.png | 180.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6794 http://ftp.pdbj.org/pub/emdb/structures/EMD-6794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6794 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6794.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6794.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | zika virus complexed with Fab of a human monoclonal antibody named ZKA190 at 4 celsius degree | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : zika virus complexed with Fab molecule of a human monoclonal anti...
| Entire | Name: zika virus complexed with Fab molecule of a human monoclonal antibody named ZKA190 |
|---|---|
| Components |
|
-Supramolecule #1: zika virus complexed with Fab molecule of a human monoclonal anti...
| Supramolecule | Name: zika virus complexed with Fab molecule of a human monoclonal antibody named ZKA190 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:    Zika virus / Strain: H/PF/2013 Zika virus / Strain: H/PF/2013 |
-Supramolecule #2: zika virus
| Supramolecule | Name: zika virus / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:    Zika virus / Strain: H/PF/2013 Zika virus / Strain: H/PF/2013 |
-Supramolecule #3: Fab ZKA190
| Supramolecule | Name: Fab ZKA190 / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: EXPI293 Homo sapiens (human) / Recombinant cell: EXPI293 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||
| Grid | Material: COPPER / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm Bright-field microscopy / Cs: 2.7 mm |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller