+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5220 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The architecture of the DNA polymerase-PCNA-DNA ternary complex | |||||||||
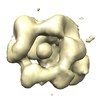
 Map data Map data | This is a map of PolB-PCNA-DNA complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  DNA replication / DNA replication /  DNA clamp DNA clamp | |||||||||
| Biological species |    Pyrococcus furiosus (archaea) / unidentified (others) Pyrococcus furiosus (archaea) / unidentified (others) | |||||||||
| Method |  single particle reconstruction / Resolution: 19.0 Å single particle reconstruction / Resolution: 19.0 Å | |||||||||
 Authors Authors | Mayanagi K / Nishida H / Kiyonari S / Saito M / Kohda D / Ishino Y / Shirai T / Morikawa K | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2011 Journal: Proc Natl Acad Sci U S A / Year: 2011Title: Architecture of the DNA polymerase B-proliferating cell nuclear antigen (PCNA)-DNA ternary complex. Authors: Kouta Mayanagi / Shinichi Kiyonari / Hirokazu Nishida / Mihoko Saito / Daisuke Kohda / Yoshizumi Ishino / Tsuyoshi Shirai / Kosuke Morikawa /  Abstract: DNA replication in archaea and eukaryotes is executed by family B DNA polymerases, which exhibit full activity when complexed with the DNA clamp, proliferating cell nuclear antigen (PCNA). This ...DNA replication in archaea and eukaryotes is executed by family B DNA polymerases, which exhibit full activity when complexed with the DNA clamp, proliferating cell nuclear antigen (PCNA). This replication enzyme consists of the polymerase and exonuclease moieties responsible for DNA synthesis and editing (proofreading), respectively. Because of the editing activity, this enzyme ensures the high fidelity of DNA replication. However, it remains unclear how the PCNA-complexed enzyme temporally switches between the polymerizing and editing modes. Here, we present the three-dimensional structure of the Pyrococcus furiosus DNA polymerase B-PCNA-DNA ternary complex, which is the core component of the replisome, determined by single particle electron microscopy of negatively stained samples. This structural view, representing the complex in the editing mode, revealed the whole domain configuration of the trimeric PCNA ring and the DNA polymerase, including protein-protein and protein-DNA contacts. Notably, besides the authentic DNA polymerase-PCNA interaction through a PCNA-interacting protein (PIP) box, a novel contact was found between DNA polymerase and the PCNA subunit adjacent to that with the PIP contact. This contact appears to be responsible for the configuration of the complex specific for the editing mode. The DNA was located almost at the center of PCNA and exhibited a substantial and particular tilt angle against the PCNA ring plane. The obtained molecular architecture of the complex, including the new contact found in this work, provides clearer insights into the switching mechanism between the two distinct modes, thus highlighting the functional significance of PCNA in the replication process. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5220.map.gz emd_5220.map.gz | 217.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5220-v30.xml emd-5220-v30.xml emd-5220.xml emd-5220.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5220_1.jpg emd_5220_1.jpg | 29.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5220 http://ftp.pdbj.org/pub/emdb/structures/EMD-5220 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5220 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5220 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5220.map.gz / Format: CCP4 / Size: 670.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5220.map.gz / Format: CCP4 / Size: 670.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of PolB-PCNA-DNA complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pyrococcus furiosus PolB-PCNA-DNA
| Entire | Name: Pyrococcus furiosus PolB-PCNA-DNA |
|---|---|
| Components |
|
-Supramolecule #1000: Pyrococcus furiosus PolB-PCNA-DNA
| Supramolecule | Name: Pyrococcus furiosus PolB-PCNA-DNA / type: sample / ID: 1000 / Details: The sample was monodisperse Oligomeric state: One PolB binds to one trimer of PCNA and primed DNA Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 210 KDa |
-Macromolecule #1: DNA polymerase
| Macromolecule | Name: DNA polymerase / type: protein_or_peptide / ID: 1 / Name.synonym: PolB / Oligomeric state: monomer / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:    Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Theoretical: 104 KDa |
-Macromolecule #2: proliferating cell nuclear antigen
| Macromolecule | Name: proliferating cell nuclear antigen / type: protein_or_peptide / ID: 2 / Name.synonym: PCNA / Oligomeric state: trimer / Recombinant expression: Yes / Database: NCBI |
|---|---|
| Source (natural) | Organism:    Pyrococcus furiosus (archaea) Pyrococcus furiosus (archaea) |
| Molecular weight | Theoretical: 104 KDa |
-Macromolecule #3: DNA
| Macromolecule | Name: DNA / type: dna / ID: 3 / Name.synonym: DNA / Classification: DNA / Structure: DOUBLE HELIX / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism: unidentified (others) |
| Molecular weight | Theoretical: 23 KDa |
-Experimental details
-Structure determination
 Processing Processing |  single particle reconstruction single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 50 mM Tris-HCl,5 mM MgCl2 |
| Grid | Details: 200 mesh Cu grid with carbon support film |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1010 |
|---|---|
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 400000 Bright-field microscopy / Nominal magnification: 400000 |
| Sample stage | Specimen holder: Side entry standard / Specimen holder model: JEOL |
| Alignment procedure | Legacy - Astigmatism: objective astigmatism was corrected using a quadrupole stigmator at 400,000 times magnification. |
| Details | minimum dose |
| Image recording | Category: CCD / Film or detector model: GENERIC GATAN |
- Image processing
Image processing
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 19.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN, IMAGIC / Number images used: 18897 |
|---|
 Movie
Movie Controller
Controller