[English] 日本語
 Yorodumi
Yorodumi- EMDB-3754: The cryo-EM structure of Tick-borne encephalitis virus complexed ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3754 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of Tick-borne encephalitis virus complexed with Fab fragment of neutralizing antibody 19/1786 | ||||||||||||
 Map data Map data | Tick-borne encephalitis virus in complex with Fab fragment of neutralizing antibody 19/1786 | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology information flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity /  viral capsid / nucleoside-triphosphate phosphatase / viral capsid / nucleoside-triphosphate phosphatase /  double-stranded RNA binding / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / clathrin-dependent endocytosis of virus by host cell / double-stranded RNA binding / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / clathrin-dependent endocytosis of virus by host cell /  mRNA (nucleoside-2'-O-)-methyltransferase activity ... mRNA (nucleoside-2'-O-)-methyltransferase activity ... flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity /  viral capsid / nucleoside-triphosphate phosphatase / viral capsid / nucleoside-triphosphate phosphatase /  double-stranded RNA binding / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / clathrin-dependent endocytosis of virus by host cell / double-stranded RNA binding / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / clathrin-dependent endocytosis of virus by host cell /  mRNA (nucleoside-2'-O-)-methyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / mRNA (nucleoside-2'-O-)-methyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity /  RNA helicase activity / host cell endoplasmic reticulum membrane / host cell perinuclear region of cytoplasm / RNA helicase activity / host cell endoplasmic reticulum membrane / host cell perinuclear region of cytoplasm /  protein dimerization activity / protein dimerization activity /  RNA helicase / induction by virus of host autophagy / RNA helicase / induction by virus of host autophagy /  RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase / viral RNA genome replication /  RNA-dependent RNA polymerase activity / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / RNA-dependent RNA polymerase activity / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane /  viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / host cell nucleus / structural molecule activity / virion attachment to host cell / virion membrane / viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / host cell nucleus / structural molecule activity / virion attachment to host cell / virion membrane /  ATP hydrolysis activity / ATP hydrolysis activity /  proteolysis / extracellular region / proteolysis / extracellular region /  ATP binding / ATP binding /  membrane / membrane /  metal ion binding metal ion bindingSimilarity search - Function | ||||||||||||
| Biological species |   Mus musculus (house mouse) / Mus musculus (house mouse) /   Tick-borne encephalitis virus (strain Hypr) / Tick-borne encephalitis virus (strain Hypr) /   House mouse (house mouse) / House mouse (house mouse) /   Tick-borne encephalitis virus (strain HYPR) Tick-borne encephalitis virus (strain HYPR) | ||||||||||||
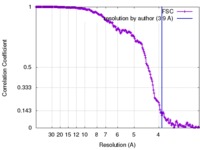
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.9 Å cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Fuzik T / Plevka P | ||||||||||||
| Funding support |  Czech Republic, Czech Republic,  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Structure of tick-borne encephalitis virus and its neutralization by a monoclonal antibody. Authors: Tibor Füzik / Petra Formanová / Daniel Růžek / Kentaro Yoshii / Matthias Niedrig / Pavel Plevka /    Abstract: Tick-borne encephalitis virus (TBEV) causes 13,000 cases of human meningitis and encephalitis annually. However, the structure of the TBEV virion and its interactions with antibodies are unknown. ...Tick-borne encephalitis virus (TBEV) causes 13,000 cases of human meningitis and encephalitis annually. However, the structure of the TBEV virion and its interactions with antibodies are unknown. Here, we present cryo-EM structures of the native TBEV virion and its complex with Fab fragments of neutralizing antibody 19/1786. Flavivirus genome delivery depends on membrane fusion that is triggered at low pH. The virion structure indicates that the repulsive interactions of histidine side chains, which become protonated at low pH, may contribute to the disruption of heterotetramers of the TBEV envelope and membrane proteins and induce detachment of the envelope protein ectodomains from the virus membrane. The Fab fragments bind to 120 out of the 180 envelope glycoproteins of the TBEV virion. Unlike most of the previously studied flavivirus-neutralizing antibodies, the Fab fragments do not lock the E-proteins in the native-like arrangement, but interfere with the process of virus-induced membrane fusion. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3754.map.gz emd_3754.map.gz | 70.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3754-v30.xml emd-3754-v30.xml emd-3754.xml emd-3754.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3754_fsc.xml emd_3754_fsc.xml | 17.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_3754.png emd_3754.png | 322.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3754 http://ftp.pdbj.org/pub/emdb/structures/EMD-3754 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3754 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3754 | HTTPS FTP |
-Related structure data
| Related structure data |  5o6vMC  3752C  3755C  5o6aC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3754.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3754.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tick-borne encephalitis virus in complex with Fab fragment of neutralizing antibody 19/1786 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.45 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tick-borne encephalitis virus (strain HYPR) and Fab
| Entire | Name: Tick-borne encephalitis virus (strain HYPR) and Fab |
|---|---|
| Components |
|
-Supramolecule #1: Tick-borne encephalitis virus (strain HYPR) and Fab
| Supramolecule | Name: Tick-borne encephalitis virus (strain HYPR) and Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #3: Mus musculus Fab
| Supramolecule | Name: Mus musculus Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Supramolecule #2: Tick-borne encephalitis virus (strain HYPR)
| Supramolecule | Name: Tick-borne encephalitis virus (strain HYPR) / type: virus / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 / NCBI-ID: 70733 Sci species name: Tick-borne encephalitis virus (strain HYPR) Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Envelope protein
| Macromolecule | Name: Envelope protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tick-borne encephalitis virus (strain Hypr) Tick-borne encephalitis virus (strain Hypr) |
| Molecular weight | Theoretical: 53.667418 KDa |
| Sequence | String: SRCTHLENRD FVTGTQGTTR VTLVLELGGC VTITAEGKPS MDVWLDAIYQ ENPAQTREYC LHAKLSDTKV AARCPTMGPA TLAEEHQGG TVCKRDQSDR GWGNHCGLFG KGSIVACVKA ACEAKKKATG HVYDANKIVY TVKVEPHTGD YVAANETHSG R KTASFTVS ...String: SRCTHLENRD FVTGTQGTTR VTLVLELGGC VTITAEGKPS MDVWLDAIYQ ENPAQTREYC LHAKLSDTKV AARCPTMGPA TLAEEHQGG TVCKRDQSDR GWGNHCGLFG KGSIVACVKA ACEAKKKATG HVYDANKIVY TVKVEPHTGD YVAANETHSG R KTASFTVS SEKTILTMGE YGDVSLLCRV ASGVDLAQTV ILELDKTVEH LPTAWQVHRD WFNDLALPWK HEGARNWNNA ER LVEFGAP HAVKMDVYNL GDQTGVLLKA LAGVPVAHIE GTKYHLKSGH VTCEVGLEKL KMKGLTYTMC DKTKFTWKRA PTD SGHDTV VMEVTFSGTK PCRIPVRAVA HGSPDVNVAM LITPNPTIEN NGGGFIEMQL PPGDNIIYVG ELSYQWFQKG SSIG RVFQK TKKGIERLTV IGEHAWDFGS AGGFLSSIGK ALHTVLGGAF NSIFGGVGFL PKLLLGVALA WLGLNMRNPT MSMSF LLAG VLVLAMTLGV GA |
-Macromolecule #2: Small envelope protein M
| Macromolecule | Name: Small envelope protein M / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Tick-borne encephalitis virus (strain Hypr) Tick-borne encephalitis virus (strain Hypr) |
| Molecular weight | Theoretical: 8.339867 KDa |
| Sequence | String: SVLIPSHAQG ELTGRGHKWL EGDSLRTHLT RVEGWVWKNR LLALAMVTVV WLTLESVVTR VAVLVVLLCL APVYA |
-Macromolecule #3: Fab 19/1786 - Heavy chain
| Macromolecule | Name: Fab 19/1786 - Heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   House mouse (house mouse) House mouse (house mouse) |
| Molecular weight | Theoretical: 22.730357 KDa |
| Sequence | String: EVKLEESGGG LVQPGGSLKL SCAASGFTFS SYGMSWVRQT PDKRLELVAA TNSDGDSTYY PDSVKGRFTI SRDRAKNTLY LQMSSLKSE DTAMYYCTRV LYDYDGEFAY WGQGTLVTVS AAKTTPPSVY PLAPGSAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS ...String: EVKLEESGGG LVQPGGSLKL SCAASGFTFS SYGMSWVRQT PDKRLELVAA TNSDGDSTYY PDSVKGRFTI SRDRAKNTLY LQMSSLKSE DTAMYYCTRV LYDYDGEFAY WGQGTLVTVS AAKTTPPSVY PLAPGSAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS GVHTFPAVLQ SDLYTLSSSV TVPSKTWPSE TVTCNVAHPA SSTK |
-Macromolecule #4: Fab 19/1786 - Light chain
| Macromolecule | Name: Fab 19/1786 - Light chain / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   House mouse (house mouse) House mouse (house mouse) |
| Molecular weight | Theoretical: 22.84718 KDa |
| Sequence | String: DIVMTQSQKF MSTSVGDRVT VTCKASQNVG TNVAWYQQKP GQSPKGLIYS ASYRYSGVPD RFIGSGSGTD FTLTISNVQS GDLAEYFCQ QYNNHPLTFG AGTKLELKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQDGV L NSWTDQDS ...String: DIVMTQSQKF MSTSVGDRVT VTCKASQNVG TNVAWYQQKP GQSPKGLIYS ASYRYSGVPD RFIGSGSGTD FTLTISNVQS GDLAEYFCQ QYNNHPLTFG AGTKLELKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQDGV L NSWTDQDS KDSTYSMSST LTLTKDEYER HNSYTCEATH KTSTSPIVKS |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: NITROGEN / Pretreatment - Pressure: 0.007 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: Wait time: 10 s Blot time: 2 s Blot force: -2 Drain time: 0 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 75000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 75000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Digitization - Frames/image: 1-7 / Number grids imaged: 1 / Number real images: 5426 / Average exposure time: 0.5 sec. / Average electron dose: 22.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Details | An initial model was generated based on the tick-borne encephalitis virus (TBEV) cryo-EM structure (PDB:5O6A) and a homology modle of the Fab 19/1786. The model was rigid-body fitted to the electron density map of the TBEV+Fab 19/1786 using the program Chimera. Subsequently, the structure was manually corrected using the program Coot, followed by real-space refinement in Phenix and reciprocal space refinement in Refmac5. |
|---|---|
| Refinement | Space: RECIPROCAL / Protocol: OTHER |
| Output model |  PDB-5o6v: |
 Movie
Movie Controller
Controller