[English] 日本語
 Yorodumi
Yorodumi- EMDB-3601: Cryo EM structure of the conjugative relaxase TraI of the F/R1 pl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3601 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo EM structure of the conjugative relaxase TraI of the F/R1 plasmid system | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information DNA topoisomerase / DNA topoisomerase type I (single strand cut, ATP-independent) activity / hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides / DNA topoisomerase / DNA topoisomerase type I (single strand cut, ATP-independent) activity / hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides /  metabolic process / metabolic process /  DNA helicase activity / DNA helicase activity /  DNA helicase / DNA helicase /  ATP hydrolysis activity / ATP hydrolysis activity /  DNA binding / DNA binding /  ATP binding / ATP binding /  metal ion binding / metal ion binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||
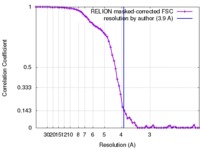
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.9 Å cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Zanetti G / Ilangovan A / Waksman G | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: Cryo-EM Structure of a Relaxase Reveals the Molecular Basis of DNA Unwinding during Bacterial Conjugation. Authors: Aravindan Ilangovan / Christopher W M Kay / Sandro Roier / Hassane El Mkami / Enrico Salvadori / Ellen L Zechner / Giulia Zanetti / Gabriel Waksman /   Abstract: Relaxases play essential roles in conjugation, the main process by which bacteria exchange genetic material, notably antibiotic resistance genes. They are bifunctional enzymes containing a trans- ...Relaxases play essential roles in conjugation, the main process by which bacteria exchange genetic material, notably antibiotic resistance genes. They are bifunctional enzymes containing a trans-esterase activity, which is responsible for nicking the DNA strand to be transferred and for covalent attachment to the resulting 5'-phosphate end, and a helicase activity, which is responsible for unwinding the DNA while it is being transported to a recipient cell. Here we show that these two activities are carried out by two conformers that can both load simultaneously on the origin of transfer DNA. We solve the structure of one of these conformers by cryo electron microscopy to near-atomic resolution, elucidating the molecular basis of helicase function by relaxases and revealing insights into the mechanistic events taking place in the cell prior to substrate transport during conjugation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3601.map.gz emd_3601.map.gz | 25.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3601-v30.xml emd-3601-v30.xml emd-3601.xml emd-3601.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3601_fsc.xml emd_3601_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_3601.png emd_3601.png | 155.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3601 http://ftp.pdbj.org/pub/emdb/structures/EMD-3601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3601 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3601 | HTTPS FTP |
-Related structure data
| Related structure data |  5n8oMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3601.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3601.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TraI-22mer complex
| Entire | Name: TraI-22mer complex |
|---|---|
| Components |
|
-Supramolecule #1: TraI-22mer complex
| Supramolecule | Name: TraI-22mer complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Experimental: 193 KDa |
-Supramolecule #2: TraI protein
| Supramolecule | Name: TraI protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Supramolecule #3: 22-mer DNA/RNA hybrid
| Supramolecule | Name: 22-mer DNA/RNA hybrid / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism: synthetic construct (others) |
-Macromolecule #1: DNA helicase I
| Macromolecule | Name: DNA helicase I / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 191.996734 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MMSIAQVRSA GSAGNYYTDK DNYYVLGSMG ERWAGKGAEQ LGLQGSVDKD VFTRLLEGRL PDGADLSRMQ DGSNKHRPGY DLTFSAPKS VSMMAMLGGD KRLIDAHNQA VDFAVRQVEA LASTRVMTDG QSETVLTGNL VMALFNHDTS RDQEPQLHTH A VVANVTQH ...String: MMSIAQVRSA GSAGNYYTDK DNYYVLGSMG ERWAGKGAEQ LGLQGSVDKD VFTRLLEGRL PDGADLSRMQ DGSNKHRPGY DLTFSAPKS VSMMAMLGGD KRLIDAHNQA VDFAVRQVEA LASTRVMTDG QSETVLTGNL VMALFNHDTS RDQEPQLHTH A VVANVTQH NGEWKTLSSD KVGKTGFIEN VYANQIAFGR LYREKLKEQV EALGYETEVV GKHGMWEMPG VPVEAFSGRS QA IREAVGE DASLKSRDVA ALDTRKSKQH VDPEIRMAEW MQTLKETGFD IRAYRDAADQ RTEIRTQAPG PASQDGPDVQ QAV TQAIAG LSERKVQFTY TDVLARTVGI LPPENGVIER ARAGIDEAIS REQLIPLDRE KGLFTSGIHV LDELSVRALS RDIM KQNRV TVHPEKSVPR TAGYSDAVSV LAQDRPSLAI VSGQGGAAGQ RERVAELVMM AREQGREVQI IAADRRSQMN LKQDE RLSG ELITGRRQLL EGMAFTPGST VIVDQGEKLS LKETLTLLDG AARHNVQVLI TDSGQRTGTG SALMAMKDAG VNTYRW QGG EQRPATIISE PDRNVRYARL AGDFAASVKA GEESVAQVSG VREQAILTQA IRSELKTQGV LGHPEVTMTA LSPVWLD SR SRYLRDMYRP GMVMEQWNPE TRSHDRYVID RVTAQSHSLT LRDAQGETQV VRISSLDSSW SLFRPEKMPV ADGERLRV T GKIPGLRVSG GDRLQVASVS EDAMTVVVPG RAEPASLPVS DSPFTALKLE NGWVETPGHS VSDSATVFAS VTQMAMDNA TLNGLARSGR DVRLYSSLDE TRTAEKLARH PSFTVVSEQI KARAGETLLE TAISLQKAGL HTPAQQAIHL ALPVLESKNL AFSMVDLLT EAKSFAAEGT GFTELGGEIN AQIKRGDLLY VDVAKGYGTG LLVSRASYEA EKSILRHILE GKEAVTPLME R VPGELMET LTSGQRAATR MILETSDRFT VVQGYAGVGK TTQFRAVMSA VNMLPASERP RVVGLGPTHR AVGEMRSAGV DA QTLASFL HDTQLQQRSG ETPDFSNTLF LLDESSMVGN TEMARAYALI AAGGGRAVAS GDTDQLQAIA PGQSFRLQQT RSA ADVVIM KEIVRQTPEL REAVYSLINR DVERALSGLE SVKPSQVPRL EGAWAPEHSV TEFSHSQEAK LAEAQQKAML KGEA FPDIP MTLYEAIVRD YTGRTPEARE QTLIVTHLNE DRRVLNSMIH DAREKAGELG KEQVMVPVLN TANIRDGELR RLSTW EKNP DALALVDNVY HRIAGISKDD GLITLQDAEG NTRLISPREA VAEGVTLYTP DKIRVGTGDR MRFTKSDRER GYVANS VWT VTAVSGDSVT LSDGQQTRVI RPGQERAEQH IDLAYAITAH GAQGASETFA IALEGTEGNR KLMAGFESAY VALSRMK QH VQVYTDNRQG WTDAINNAVQ KGTAHDVLEP KPDREVMNAQ RLFSTARELR DVAAGRAVLR QAGLAGGDSP ARFIAPGR K YPQPYVALPA FDRNGKSAGI WLNPLTTDDG NGLRGFSGEG RVKGSGDAQF VALQGSRNGE SLLADNMQDG VRIARDNPD SGVVVRIAGE GRPWNPGAIT GGRVWGDIPD NSVQPGAGNG EPVTAEVLAQ RQAEEAIRRE TERRADEIVR KMAENKPDLP DGKTELAVR DIAGQERDRS AISERETALP ESVLRESQRE REAVREVARE NLLQERLQQM ERDMVRDLQK EKTLGGD |
-Macromolecule #2: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3') type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 6.647284 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK III / Details: blot time 4 sec force 1. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 47619 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Specialist optics | Energy filter - Name: GIF / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-20 / Number grids imaged: 1 / Number real images: 2900 / Average exposure time: 0.4 sec. / Average electron dose: 2.5 e/Å2 / Details: Total exposure 8 sec for a total dose of 50 e- |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Details | Please see article for details of model building and refinement |
|---|---|
| Refinement | Space: REAL |
| Output model |  PDB-5n8o: |
 Movie
Movie Controller
Controller