+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3417 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of tetrameric MotA complex | |||||||||
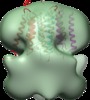
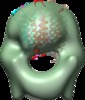
 Map data Map data | Reconstruction of Aquifex aeolicus stator protein, tetrameric MotA complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Flagella motor /  Stator / Stator /  Single Particle analysis / Single Particle analysis /  Proton motive force Proton motive force | |||||||||
| Function / homology |  Function and homology information Function and homology informationbacterial-type flagellum-dependent swarming motility / proton transmembrane transport /  chemotaxis / membrane => GO:0016020 / chemotaxis / membrane => GO:0016020 /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |    Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 25.0 Å negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Takekawa N / Terahara N / Kato T / Gohara M / Mayanagi K / Hijikata A / Onoue Y / Kojima S / Shirai T / Namba K / Homma M | |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2016 Journal: Sci Rep / Year: 2016Title: The tetrameric MotA complex as the core of the flagellar motor stator from hyperthermophilic bacterium. Authors: Norihiro Takekawa / Naoya Terahara / Takayuki Kato / Mizuki Gohara / Kouta Mayanagi / Atsushi Hijikata / Yasuhiro Onoue / Seiji Kojima / Tsuyoshi Shirai / Keiichi Namba / Michio Homma /  Abstract: Rotation of bacterial flagellar motor is driven by the interaction between the stator and rotor, and the driving energy is supplied by ion influx through the stator channel. The stator is composed of ...Rotation of bacterial flagellar motor is driven by the interaction between the stator and rotor, and the driving energy is supplied by ion influx through the stator channel. The stator is composed of the MotA and MotB proteins, which form a hetero-hexameric complex with a stoichiometry of four MotA and two MotB molecules. MotA and MotB are four- and single-transmembrane proteins, respectively. To generate torque, the MotA/MotB stator unit changes its conformation in response to the ion influx, and interacts with the rotor protein FliG. Here, we overproduced and purified MotA of the hyperthermophilic bacterium Aquifex aeolicus. A chemical crosslinking experiment revealed that MotA formed a multimeric complex, most likely a tetramer. The three-dimensional structure of the purified MotA, reconstructed by electron microscopy single particle imaging, consisted of a slightly elongated globular domain and a pair of arch-like domains with spiky projections, likely to correspond to the transmembrane and cytoplasmic domains, respectively. We show that MotA molecules can form a stable tetrameric complex without MotB, and for the first time, demonstrate the cytoplasmic structure of the stator. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3417.map.gz emd_3417.map.gz | 741.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3417-v30.xml emd-3417-v30.xml emd-3417.xml emd-3417.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3417.png emd_3417.png emd_3417_1.png emd_3417_1.png | 327.4 KB 319.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3417 http://ftp.pdbj.org/pub/emdb/structures/EMD-3417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3417 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3417 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3417.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3417.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of Aquifex aeolicus stator protein, tetrameric MotA complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Aquifex aeolicus MotA
| Entire | Name: Aquifex aeolicus MotA |
|---|---|
| Components |
|
-Supramolecule #1000: Aquifex aeolicus MotA
| Supramolecule | Name: Aquifex aeolicus MotA / type: sample / ID: 1000 / Oligomeric state: tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 109 KDa |
-Macromolecule #1: Motility protein A
| Macromolecule | Name: Motility protein A / type: protein_or_peptide / ID: 1 / Name.synonym: Chemotaxis protein MotA / Number of copies: 4 / Oligomeric state: Tetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:    Aquifex aeolicus (bacteria) / Strain: VF5 Aquifex aeolicus (bacteria) / Strain: VF5 |
| Molecular weight | Theoretical: 109 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) / Recombinant strain: BL21-CodonPlus(DE3)-RIPL / Recombinant plasmid: pColdI Escherichia coli (E. coli) / Recombinant strain: BL21-CodonPlus(DE3)-RIPL / Recombinant plasmid: pColdI |
| Sequence | UniProtKB:  Motility protein A Motility protein A |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 50 mM Tris-HCl, 200 mM NaCl, 0.02% DMNG |
| Staining | Type: NEGATIVE / Details: 2% w/v uranyl acetate |
| Grid | Details: 200 mesh copper grid with continuous carbon suport, glow discharged |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 107140 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 4.1 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.135 µm / Nominal magnification: 80000 Bright-field microscopy / Cs: 4.1 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.135 µm / Nominal magnification: 80000 |
| Specialist optics | Energy filter - Name: Omega Filter / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Sample stage | Specimen holder: Nitrogen cooled / Specimen holder model: JEOL 3200FSC CRYOHOLDER |
| Temperature | Min: 85.5 K / Max: 85.7 K / Average: 85.5 K |
| Date | Nov 16, 2015 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Number real images: 50 / Average electron dose: 2.8 e/Å2 |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Relion-1.4 / Number images used: 5419 ) / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Relion-1.4 / Number images used: 5419 |
|---|
 Movie
Movie Controller
Controller