[English] 日本語
 Yorodumi
Yorodumi- EMDB-2339: Variable internal flexibility characterizes the helical capsid fo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2339 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
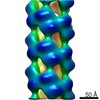
| Title | Variable internal flexibility characterizes the helical capsid formed by Agrobacterium VirE2 protein on single-stranded DNA. | |||||||||
 Map data Map data | CryoEM reconstruction of the Agrobacterium T-complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tcomplex /  agrobacterium / helical reconstruction agrobacterium / helical reconstruction | |||||||||
| Function / homology | VirE2 / VirE2 / DNA-mediated transformation / host cell nucleus /  DNA binding / extracellular region / identical protein binding / DNA binding / extracellular region / identical protein binding /  Single-strand DNA-binding protein Single-strand DNA-binding protein Function and homology information Function and homology information | |||||||||
| Biological species |  Agrobacterium fabrum str. C58 (bacteria) Agrobacterium fabrum str. C58 (bacteria) | |||||||||
| Method | helical reconstruction /  cryo EM / Resolution: 20.0 Å cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Bharat TAM / Zbaida D / Eisenstein M / Frankenstein Z / Mehlman T / Weiner L / Sorzano COS / Barak Y / Albeck S / Briggs JAG ...Bharat TAM / Zbaida D / Eisenstein M / Frankenstein Z / Mehlman T / Weiner L / Sorzano COS / Barak Y / Albeck S / Briggs JAG / Wolf SG / Elbaum M | |||||||||
 Citation Citation |  Journal: Structure / Year: 2013 Journal: Structure / Year: 2013Title: Variable internal flexibility characterizes the helical capsid formed by agrobacterium VirE2 protein on single-stranded DNA. Authors: Tanmay A M Bharat / David Zbaida / Miriam Eisenstein / Ziv Frankenstein / Tevie Mehlman / Lev Weiner / Carlos Oscar S Sorzano / Yoav Barak / Shira Albeck / John A G Briggs / Sharon G Wolf / Michael Elbaum /  Abstract: Agrobacterium is known for gene transfer to plants. In addition to a linear ssDNA oligonucleotide, Agrobacterium tumefaciens secretes an abundant ssDNA-binding effector, VirE2. In many ways VirE2 ...Agrobacterium is known for gene transfer to plants. In addition to a linear ssDNA oligonucleotide, Agrobacterium tumefaciens secretes an abundant ssDNA-binding effector, VirE2. In many ways VirE2 adapts the conjugation mechanism to transform the eukaryotic host. The crystal structure of VirE2 shows two compact domains joined by a flexible linker. Bound to ssDNA, VirE2 forms an ordered solenoidal shell, or capsid known as the T-complex. Here, we present a three-dimensional reconstruction of the VirE2-ssDNA complex using cryo-electron microscopy and iterative helical real-space reconstruction. High-resolution refinement was not possible due to inherent heterogeneity in the protein structure. By a combination of computational modeling, chemical modifications, mass spectroscopy, and electron paramagnetic resonance, we found that the N-terminal domain is tightly constrained by both tangential and longitudinal links, while the C terminus is weakly constrained. The quaternary structure is thus rigidly assembled while remaining locally flexible. This flexibility may be important in accommodating substrates without sequence specificity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2339.map.gz emd_2339.map.gz | 247.1 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2339-v30.xml emd-2339-v30.xml emd-2339.xml emd-2339.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2339.jpg emd_2339.jpg | 75.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2339 http://ftp.pdbj.org/pub/emdb/structures/EMD-2339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2339 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2339 | HTTPS FTP |
-Related structure data
| Related structure data |  4blfMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2339.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2339.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of the Agrobacterium T-complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Agrobacterium T-complex
| Entire | Name: Agrobacterium T-complex |
|---|---|
| Components |
|
-Supramolecule #1000: Agrobacterium T-complex
| Supramolecule | Name: Agrobacterium T-complex / type: sample / ID: 1000 / Oligomeric state: Helical / Number unique components: 2 |
|---|
-Macromolecule #1: VirE2
| Macromolecule | Name: VirE2 / type: protein_or_peptide / ID: 1 / Oligomeric state: Helical / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Agrobacterium fabrum str. C58 (bacteria) Agrobacterium fabrum str. C58 (bacteria) |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
-Macromolecule #2: short oligomeric 26mer DNA
| Macromolecule | Name: short oligomeric 26mer DNA / type: dna / ID: 2 / Classification: DNA / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  Agrobacterium fabrum str. C58 (bacteria) Agrobacterium fabrum str. C58 (bacteria) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 50 mM Tris, 500 mM NaCl |
| Grid | Details: Quantifoil holey carbon |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: HOMEMADE PLUNGER |
| Details | Protein was mixed with single-stranded DNA |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at high-magnification (>100,000) |
| Date | Jun 6, 2008 |
| Image recording | Category: CCD / Film or detector model: GENERIC TVIPS / Average electron dose: 20 e/Å2 / Details: Image data was collected as focal pairs. |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Phase-flipping |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 14.67 Å Applied symmetry - Helical parameters - Δ&Phi: 110.09 ° Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Bsoft, EMAN, Xmipp, Spider, IHRSR |
| Details | Particles were picked and preselected using routines of Xmipp, and then reconstruction was carried out using IHRSR. |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: fitPDB2EM |
| Details | The N and C terminal domain were fit separately by exhaustive molecular modeling using the fitPDB2EM program. Only the N-terminal domain could be constrained strongly. |
| Refinement | Space: RECIPROCAL / Protocol: FLEXIBLE FIT / Target criteria: Highest cross-correlation |
| Output model |  PDB-4blf: |
 Movie
Movie Controller
Controller