Yorodumi
Yorodumi+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1534 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EcoKI type I RM methyltransferase with DNA mimic Ocr. Negative stain 3D. | |||||||||
 Map data Map data | Negative stain EM reconstruction of M.EcoKI-Ocr (Two different models submitted to PDB: 2Y7C, 2Y7H) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | EcoKI /  methyltransferase / type I restriction / Ocr / methyltransferase / type I restriction / Ocr /  HsdS / HsdM / T7 HsdS / HsdM / T7 | |||||||||
| Function / homology |  Function and homology information Function and homology information type I site-specific deoxyribonuclease complex / symbiont-mediated evasion of host restriction-modification system / type I site-specific deoxyribonuclease complex / symbiont-mediated evasion of host restriction-modification system /  protein binding / N-methyltransferase activity / protein binding / N-methyltransferase activity /  site-specific DNA-methyltransferase (adenine-specific) / site-specific DNA-methyltransferase (adenine-specific) /  site-specific DNA-methyltransferase (adenine-specific) activity / DNA restriction-modification system / site-specific DNA-methyltransferase (adenine-specific) activity / DNA restriction-modification system /  DNA binding / DNA binding /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli (E. coli) / Escherichia coli (E. coli) /    Enterobacteria phage T7 (virus) Enterobacteria phage T7 (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 18.0 Å negative staining / Resolution: 18.0 Å | |||||||||
 Authors Authors | Kennaway CK / Obarska-Kosinska A / White JH / Tuszynska I / Cooper LP / Bujnicki JM / Trinick J / Dryden DTF | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2009 Journal: Nucleic Acids Res / Year: 2009Title: The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Authors: Christopher K Kennaway / Agnieszka Obarska-Kosinska / John H White / Irina Tuszynska / Laurie P Cooper / Janusz M Bujnicki / John Trinick / David T F Dryden /  Abstract: Type-I DNA restriction-modification (R/M) systems are important agents in limiting the transmission of mobile genetic elements responsible for spreading bacterial resistance to antibiotics. EcoKI, a ...Type-I DNA restriction-modification (R/M) systems are important agents in limiting the transmission of mobile genetic elements responsible for spreading bacterial resistance to antibiotics. EcoKI, a Type I R/M enzyme from Escherichia coli, acts by methylation- and sequence-specific recognition, leading to either methylation of DNA or translocation and cutting at a random site, often hundreds of base pairs away. Consisting of one specificity subunit, two modification subunits, and two DNA translocase/endonuclease subunits, EcoKI is inhibited by the T7 phage antirestriction protein ocr, a DNA mimic. We present a 3D density map generated by negative-stain electron microscopy and single particle analysis of the central core of the restriction complex, the M.EcoKI M(2)S(1) methyltransferase, bound to ocr. We also present complete atomic models of M.EcoKI in complex with ocr and its cognate DNA giving a clear picture of the overall clamp-like operation of the enzyme. The model is consistent with a large body of experimental data on EcoKI published over 40 years. | |||||||||
| History |
|
- Structure visualization
Structure visualization
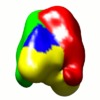
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1534.map.gz emd_1534.map.gz | 329.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1534-v30.xml emd-1534-v30.xml emd-1534.xml emd-1534.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  1534.gif 1534.gif | 38.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1534 http://ftp.pdbj.org/pub/emdb/structures/EMD-1534 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1534 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1534 | HTTPS FTP |
-Related structure data
| Related structure data |  2y7cMC  2y7hMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1534.map.gz / Format: CCP4 / Size: 422.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1534.map.gz / Format: CCP4 / Size: 422.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain EM reconstruction of M.EcoKI-Ocr (Two different models submitted to PDB: 2Y7C, 2Y7H) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.125 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
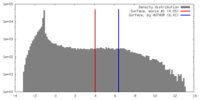
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E. coli M.EcoKI (M2S1) bound to dimeric ocr from T7 phage
| Entire | Name: E. coli M.EcoKI (M2S1) bound to dimeric ocr from T7 phage |
|---|---|
| Components |
|
-Supramolecule #1000: E. coli M.EcoKI (M2S1) bound to dimeric ocr from T7 phage
| Supramolecule | Name: E. coli M.EcoKI (M2S1) bound to dimeric ocr from T7 phage type: sample / ID: 1000 / Details: stained with 1pc uranyl acetate / Oligomeric state: HsdS-(HsdM)2-(ocr)2 / Number unique components: 5 |
|---|---|
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: HsdS
| Macromolecule | Name: HsdS / type: protein_or_peptide / ID: 1 / Name.synonym: EcoKI specificity subunit / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) / Strain: K-12 / Location in cell: Cytoplasmic Escherichia coli (E. coli) / Strain: K-12 / Location in cell: Cytoplasmic |
| Molecular weight | Theoretical: 51.4 KDa |
| Sequence | GO:  protein binding protein bindingInterPro: Type I restriction modification DNA specificity domain |
-Macromolecule #2: HsdM
| Macromolecule | Name: HsdM / type: protein_or_peptide / ID: 2 / Name.synonym: EcoKI methylase subunit / Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) / Strain: K-12 / Location in cell: cytoplasm Escherichia coli (E. coli) / Strain: K-12 / Location in cell: cytoplasm |
| Molecular weight | Theoretical: 59.3 KDa |
| Sequence | GO:  protein binding / InterPro: DNA methylase, adenine-specific protein binding / InterPro: DNA methylase, adenine-specific |
-Macromolecule #3: 0.3 gene
| Macromolecule | Name: 0.3 gene / type: protein_or_peptide / ID: 3 / Name.synonym: ocr, 0.3 gene, from T7 phage / Number of copies: 2 / Oligomeric state: Dimer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:    Enterobacteria phage T7 (virus) / synonym: phage T7 Enterobacteria phage T7 (virus) / synonym: phage T7 |
| Molecular weight | Theoretical: 13.8 KDa |
| Sequence | InterPro: Protein Ocr |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.050 mg/mL |
|---|---|
| Buffer | pH: 4.7 / Details: 20mM Tris-Cl, 100 mM NaCl, |
| Staining | Type: NEGATIVE Details: Protein was adsorbed onto UV treated carbon for 2 mins, blotted, then 1% uranyl acetate solution was applied for 1 min then blotted, twice. |
| Grid | Details: 400 mesh copper |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy #1
Electron microscopy #1
| Microscope | JEOL 1200EX |
|---|---|
| Electron beam | Acceleration voltage: 80 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 0.87 µm / Nominal defocus min: 0.275 µm / Nominal magnification: 50000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 0.87 µm / Nominal defocus min: 0.275 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side entry / Specimen holder model: OTHER |
| Temperature | Average: 294 K |
| Microscopy ID | 1 |
| Alignment procedure | Legacy - Astigmatism: Corrected at 80,000x |
| Details | Customised JEOL 1200 EX microscope, low dose mode. |
| Date | Feb 1, 2008 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.25 µm / Number real images: 12 / Average electron dose: 25 e/Å2 / Bits/pixel: 14 |
- Electron microscopy #2
Electron microscopy #2
| Microscope | JEOL 1200EX |
|---|---|
| Electron beam | Acceleration voltage: 80 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 0.87 µm / Nominal defocus min: 0.275 µm / Nominal magnification: 40000 Bright-field microscopy / Cs: 2.0 mm / Nominal defocus max: 0.87 µm / Nominal defocus min: 0.275 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder: Side entry / Specimen holder model: OTHER |
| Temperature | Average: 294 K |
| Microscopy ID | 2 |
| Alignment procedure | Legacy - Astigmatism: Corrected at 80,000x |
| Details | Customised JEOL 1200 EX microscope, low dose mode. |
| Date | Feb 1, 2008 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.25 µm / Number real images: 12 / Average electron dose: 25 e/Å2 / Bits/pixel: 14 |
- Image processing
Image processing
| CTF correction | Details: Filtered at 1st zero |
|---|---|
| Final two d classification | Number classes: 600 |
| Final angle assignment | Details: EMAN |
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN, IMAGIC ) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 18.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN, IMAGICDetails: Final map calculated from combined datasets taken at 50,000x and 40,000. Number images used: 17807 |
| Details | The particles were selected using boxer in autobox mode. |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: Situs |
| Details | Protocol: Rigid body. atomic model of whole complex (M2,S1,ocr2) fitted as single rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-2y7c:  PDB-2y7h: |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B |
|---|---|
| Software | Name: Situs |
| Details | Protocol: Rigid body. HsdS homology model based on 1yf2 used. Atomic model of whole complex (M2,S1,ocr2) fitted as single rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-2y7c:  PDB-2y7h: |
-Atomic model buiding 3
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D / Chain - #4 - Chain ID: E / Chain - #5 - Chain ID: F / Chain - #6 - Chain ID: G / Chain - #7 - Chain ID: H |
|---|---|
| Software | Name: Situs |
| Details | Protocol: Rigid body. Modified version of hsdM pdb used with alternative chain trace. Atomic model of whole complex (M2,S1,ocr2) fitted as single rigid body |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-2y7c:  PDB-2y7h: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)