+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9400 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
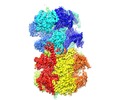
| Title | Cryo-EM structure of Mcm2-7 double hexamer on dsDNA | |||||||||
 Map data Map data | Mcm2-7 double hexamer on dsDNA | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationMCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / nuclear DNA replication / MCM complex binding / premeiotic DNA replication / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / mitotic DNA replication / Activation of the pre-replicative complex / CMG complex ...MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / nuclear DNA replication / MCM complex binding / premeiotic DNA replication / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / mitotic DNA replication / Activation of the pre-replicative complex / CMG complex / nuclear pre-replicative complex / Activation of ATR in response to replication stress / MCM complex / DNA replication preinitiation complex / double-strand break repair via break-induced replication / single-stranded DNA helicase activity / replication fork protection complex / mitotic DNA replication initiation / silent mating-type cassette heterochromatin formation / regulation of DNA-templated DNA replication initiation / DNA strand elongation involved in DNA replication / DNA unwinding involved in DNA replication / nuclear replication fork /  DNA replication origin binding / subtelomeric heterochromatin formation / DNA replication initiation / heterochromatin formation / DNA replication origin binding / subtelomeric heterochromatin formation / DNA replication initiation / heterochromatin formation /  DNA helicase activity / DNA helicase activity /  helicase activity / helicase activity /  single-stranded DNA binding / single-stranded DNA binding /  DNA helicase / DNA helicase /  chromosome, telomeric region / DNA damage response / chromosome, telomeric region / DNA damage response /  chromatin binding / chromatin binding /  ATP hydrolysis activity / ATP hydrolysis activity /  nucleoplasm / nucleoplasm /  ATP binding / ATP binding /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.9 Å cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Li H / Yuan Z / Bai L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Authors: Yasunori Noguchi / Zuanning Yuan / Lin Bai / Sarah Schneider / Gongpu Zhao / Bruce Stillman / Christian Speck / Huilin Li /   Abstract: During replication initiation, the core component of the helicase-the Mcm2-7 hexamer-is loaded on origin DNA as a double hexamer (DH). The two ring-shaped hexamers are staggered, leading to a kinked ...During replication initiation, the core component of the helicase-the Mcm2-7 hexamer-is loaded on origin DNA as a double hexamer (DH). The two ring-shaped hexamers are staggered, leading to a kinked axial channel. How the origin DNA interacts with the axial channel is not understood, but the interaction could provide key insights into Mcm2-7 function and regulation. Here, we report the cryo-EM structure of the Mcm2-7 DH on dsDNA and show that the DNA is zigzagged inside the central channel. Several of the Mcm subunit DNA-binding loops, such as the oligosaccharide-oligonucleotide loops, helix 2 insertion loops, and presensor 1 (PS1) loops, are well defined, and many of them interact extensively with the DNA. The PS1 loops of Mcm 3, 4, 6, and 7, but not 2 and 5, engage the lagging strand with an approximate step size of one base per subunit. Staggered coupling of the two opposing hexamers positions the DNA right in front of the two Mcm2-Mcm5 gates, with each strand being pressed against one gate. The architecture suggests that lagging-strand extrusion initiates in the middle of the DH that is composed of the zinc finger domains of both hexamers. To convert the Mcm2-7 DH structure into the Mcm2-7 hexamer structure found in the active helicase, the N-tier ring of the Mcm2-7 hexamer in the DH-dsDNA needs to tilt and shift laterally. We suggest that these N-tier ring movements cause the DNA strand separation and lagging-strand extrusion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9400.map.gz emd_9400.map.gz | 9.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9400-v30.xml emd-9400-v30.xml emd-9400.xml emd-9400.xml | 25.2 KB 25.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9400.png emd_9400.png | 175.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9400 http://ftp.pdbj.org/pub/emdb/structures/EMD-9400 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9400 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9400 | HTTPS FTP |
-Related structure data
| Related structure data |  5bk4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9400.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9400.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mcm2-7 double hexamer on dsDNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.088 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Mcm double hexamer bound to double-stranded DNA
+Supramolecule #1: Mcm double hexamer bound to double-stranded DNA
+Supramolecule #2: Mcm double hexamer
+Supramolecule #3: synthetic double-stranded DNA
+Macromolecule #1: DNA replication licensing factor MCM2
+Macromolecule #2: DNA replication licensing factor MCM3
+Macromolecule #3: DNA replication licensing factor MCM4
+Macromolecule #4: DNA replication licensing factor MCM5
+Macromolecule #5: DNA replication licensing factor MCM6
+Macromolecule #6: DNA replication licensing factor MCM7
+Macromolecule #7: DNA (60-mer), strand 1
+Macromolecule #8: DNA (60-mer), strand 2
+Macromolecule #9: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 10.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software: (Name: CTFFIND (ver. 4), Gctf) |
|---|---|
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 2) |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 2) |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 2) / Number images used: 58772 |
 Movie
Movie Controller
Controller