[English] 日本語
 Yorodumi
Yorodumi- EMDB-8748: Cryo-EM reconstruction of the Cafeteria roenbergensis virus capsi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8748 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
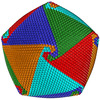
| Title | Cryo-EM reconstruction of the Cafeteria roenbergensis virus capsid suggests a novel assembly pathway for giant viruses | |||||||||
 Map data Map data | Cryo-EM reconstruction of Cafeteria roenbergensis virus with icosahedral averaging | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Cafeteria roenbergensis virus BV-PW1 Cafeteria roenbergensis virus BV-PW1 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 21.0 Å cryo EM / Resolution: 21.0 Å | |||||||||
 Authors Authors | Xiao C / Fischer MG / Bolotaulo DM / Ulloa-Rondeau N / Avila GA / Suttle CA | |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2017 Journal: Sci Rep / Year: 2017Title: Cryo-EM reconstruction of the Cafeteria roenbergensis virus capsid suggests novel assembly pathway for giant viruses. Authors: Chuan Xiao / Matthias G Fischer / Duer M Bolotaulo / Nancy Ulloa-Rondeau / Gustavo A Avila / Curtis A Suttle /    Abstract: Whereas the protein composition and overall shape of several giant virus capsids have been described, the mechanism by which these large capsids assemble remains enigmatic. Here, we present a ...Whereas the protein composition and overall shape of several giant virus capsids have been described, the mechanism by which these large capsids assemble remains enigmatic. Here, we present a reconstruction of the capsid of Cafeteria roenbergensis virus (CroV), one of the largest viruses analyzed by cryo-electron microscopy (cryo-EM) to date. The CroV capsid has a diameter of 3,000 Å and a Triangulation number of 499. Unlike related mimiviruses, the CroV capsid is not decorated with glycosylated surface fibers, but features 30 Å-long surface protrusions that are formed by loops of the major capsid protein. Based on the orientation of capsomers in the cryo-EM reconstruction, we propose that the capsids of CroV and related giant viruses are assembled by a newly conceived assembly pathway that initiates at a five-fold vertex and continuously proceeds outwards in a spiraling fashion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8748.map.gz emd_8748.map.gz | 455.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8748-v30.xml emd-8748-v30.xml emd-8748.xml emd-8748.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8748_fsc.xml emd_8748_fsc.xml | 21.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_8748.png emd_8748.png | 329.2 KB | ||
| Masks |  emd_8748_msk_1.map emd_8748_msk_1.map emd_8748_msk_2.map emd_8748_msk_2.map | 524.1 MB 524.1 MB |  Mask map Mask map | |
| Others |  emd_8748_half_map_1.map.gz emd_8748_half_map_1.map.gz emd_8748_half_map_2.map.gz emd_8748_half_map_2.map.gz | 299.8 MB 299.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8748 http://ftp.pdbj.org/pub/emdb/structures/EMD-8748 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8748 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8748 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8748.map.gz / Format: CCP4 / Size: 524.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8748.map.gz / Format: CCP4 / Size: 524.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of Cafeteria roenbergensis virus with icosahedral averaging | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.156 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
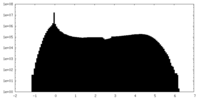
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_8748_msk_1.map emd_8748_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_8748_msk_2.map emd_8748_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Even map without soft masking of the reconstruction...
| File | emd_8748_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Even map without soft masking of the reconstruction of Cafeteria roenbergensis virus with icosahedral averaging | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Odd map without soft masking of the reconstruction...
| File | emd_8748_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Odd map without soft masking of the reconstruction of Cafeteria roenbergensis virus with icosahedral averaging | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cafeteria roenbergensis virus BV-PW1
| Entire | Name:   Cafeteria roenbergensis virus BV-PW1 Cafeteria roenbergensis virus BV-PW1 |
|---|---|
| Components |
|
-Supramolecule #1: Cafeteria roenbergensis virus BV-PW1
| Supramolecule | Name: Cafeteria roenbergensis virus BV-PW1 / type: virus / ID: 1 / Parent: 0 Details: The virus was purified following the protocol described in Fischer, M.G., et. al. The virion of Cafeteria roenbergensis virus (CroV) contains a complex suite of proteins for transcription ...Details: The virus was purified following the protocol described in Fischer, M.G., et. al. The virion of Cafeteria roenbergensis virus (CroV) contains a complex suite of proteins for transcription and DNA repair - Virology 466-467, 82-94 (2014). NCBI-ID: 693272 / Sci species name: Cafeteria roenbergensis virus BV-PW1 / Sci species strain: BV-PW1 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:   Cafeteria roenbergensis (eukaryote) Cafeteria roenbergensis (eukaryote) |
| Virus shell | Shell ID: 1 / Name: CroV / Diameter: 3000.0 Å / T number (triangulation number): 499 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Grid | Model: Quantifoil S7/2 / Material: COPPER |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/ST |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: KODAK SO-163 FILM / Average electron dose: 25.0 e/Å2 |
 Movie
Movie Controller
Controller



 Z
Z Y
Y X
X