[English] 日本語
 Yorodumi
Yorodumi- EMDB-8629: Conformational states of a soluble, uncleaved HIV-1 envelope trimer -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8629 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
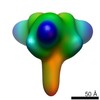
| Title | Conformational states of a soluble, uncleaved HIV-1 envelope trimer | ||||||||||||
 Map data Map data | class1 (close conformation) | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral envelope ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / positive regulation of establishment of T cell polarity / virus-mediated perturbation of host defense response / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell /  viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / structural molecule activity / virion attachment to host cell / host cell plasma membrane / virion membrane / identical protein binding / viral envelope / structural molecule activity / virion attachment to host cell / host cell plasma membrane / virion membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||
| Biological species |    Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||||||||
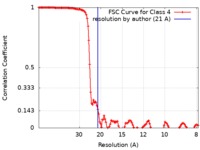
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 21.0 Å cryo EM / Resolution: 21.0 Å | ||||||||||||
 Authors Authors | Liu YH / Pan JH / Cai YF / Grigorieff N / Harrison SC / Chen B | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Virol / Year: 2017 Journal: J Virol / Year: 2017Title: Conformational States of a Soluble, Uncleaved HIV-1 Envelope Trimer. Authors: Yuhang Liu / Junhua Pan / Yongfei Cai / Nikolaus Grigorieff / Stephen C Harrison / Bing Chen /  Abstract: The HIV-1 envelope spike [Env; trimeric (gp160) cleaved to (gp120/gp41)] induces membrane fusion, leading to viral entry. It is also the viral component targeted by neutralizing antibodies. Vaccine ...The HIV-1 envelope spike [Env; trimeric (gp160) cleaved to (gp120/gp41)] induces membrane fusion, leading to viral entry. It is also the viral component targeted by neutralizing antibodies. Vaccine development requires production, in quantities suitable for clinical studies, of a recombinant form that resembles functional Env. HIV-1 gp140 trimers-the uncleaved ectodomains of (gp160)-from a few selected viral isolates adopt a compact conformation with many antigenic properties of native Env spikes. One is currently being evaluated in a clinical trial. We report here low-resolution (20 Å) electron cryomicroscopy (cryoEM) structures of this gp140 trimer, which adopts two principal conformations, one closed and the other slightly open. The former is indistinguishable at this resolution from those adopted by a stabilized, cleaved trimer (SOSIP) or by a membrane-bound Env trimer with a truncated cytoplasmic tail (EnvΔCT). The latter conformation is closer to a partially open Env trimer than to the fully open conformation induced by CD4. These results show that a stable, uncleaved HIV-1 gp140 trimer has a compact structure close to that of native Env. Development of any HIV vaccine with a protein component (for either priming or boosting) requires production of a recombinant form to mimic the trimeric, functional HIV-1 envelope spike in quantities suitable for clinical studies. Our understanding of the envelope structure has depended in part on a cleaved, soluble trimer, known as SOSIP.664, stabilized by several modifications, including an engineered disulfide. This construct, which is difficult to produce in large quantities, has yet to induce better antibody responses than those to other envelope-based immunogens, even in animal models. The uncleaved ectodomain of the envelope protein, called gp140, has also been made as a soluble form to mimic the native Env present on the virion surface. Most HIV-1 gp140 preparations are not stable, however, and have an inhomogeneous conformation. The results presented here show that gp140 preparations from suitable isolates can adopt a compact, native-like structure, supporting its use as a vaccine candidate. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8629.map.gz emd_8629.map.gz | 13.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8629-v30.xml emd-8629-v30.xml emd-8629.xml emd-8629.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8629_fsc.xml emd_8629_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_8629.png emd_8629.png | 22.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8629 http://ftp.pdbj.org/pub/emdb/structures/EMD-8629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8629 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8629 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8629.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8629.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | class1 (close conformation) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.96 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HIV-1 envelope glycoprotein gp140
| Entire | Name: HIV-1 envelope glycoprotein gp140 |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 envelope glycoprotein gp140
| Supramolecule | Name: HIV-1 envelope glycoprotein gp140 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 / Strain: C97ZA012 Human immunodeficiency virus 1 / Strain: C97ZA012 |
| Recombinant expression | Organism:   Homo sapiens (human) / Recombinant cell: HEK 293T Homo sapiens (human) / Recombinant cell: HEK 293T |
-Macromolecule #1: HIV-1 envelope glycoprotein gp140
| Macromolecule | Name: HIV-1 envelope glycoprotein gp140 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRVRGIQRNC QHLWRWGTLI LGMLMICSAA ENLWVGNMWV TVYYGVPVWT DAKTTLFCAS DTKAYDREVH NVWATHACVP TDPNPQEIVL ENVTENFNMW KNDMVDQMHE DIISLWDQSL KPCVKLTPLC VTLHCTNATF KNNVTNDMNK EIRNCSFNTT TEIRDKKQQG ...String: MRVRGIQRNC QHLWRWGTLI LGMLMICSAA ENLWVGNMWV TVYYGVPVWT DAKTTLFCAS DTKAYDREVH NVWATHACVP TDPNPQEIVL ENVTENFNMW KNDMVDQMHE DIISLWDQSL KPCVKLTPLC VTLHCTNATF KNNVTNDMNK EIRNCSFNTT TEIRDKKQQG YALFYRPDIV LLKENRNNSN NSEYILINCN ASTITQACPK VNFDPIPIHY CAPAGYAILK CNNKTFSGKG PCNNVSTVQC THGIKPVVST QLLLNGSLAE KEIIIRSENL TDNVKTIIVH LNKSVEIVCT RPNNNTRKSM RIGPGQTFYA TGDIIGDIRQ AYCNISGSKW NETLKRVKEK LQENYNNNKT IKFAPSSGGD LEITTHSFNC RGEFFYCNTT RLFNNNATED ETITLPCRIK QIINMWQGVG RAMYAPPIAG NITCKSNITG LLLVRDGGED NKTEEIFRPG GGNMKDNWRS ELYKYKVIEL KPLGIAPTGA KRRVVEREKR AVGIGAVFLG FLGAAGSTMG AASLTLTVQA RQLLSSIVQQ QSNLLRAIEA QQHMLQLTVW GIKQLQTRVL AIERYLKDQQ LLGIWGCSGK LICTTNVPWN SSWSNKSQTD IWNNMTWMEW DREISNYTDT IYRLLEDSQT QQEKNEKDLL ALDSWKNLWS WFDISNWLWY IKSRMKQIED KIEEILSKIY HIENEIARIK KLIGSGGYIP EAPRDGQAYV RKDGEWVLLS TFLGGSGRMK QIEDKIEEIE SKQKKIENEI ARIKKLGSGH HHHHH |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm Bright-field microscopy / Cs: 2.0 mm |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Sampling interval: 5.0 µm / Number grids imaged: 1 / Number real images: 3138 / Average exposure time: 8.0 sec. / Average electron dose: 40.0 e/Å2 / Details: movies were collected in super resolution mode |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller