+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8611 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain reconstruction of E. coli MCE protein YebT | ||||||||||||
 Map data Map data | E. coli MCE protein PqiB, periplasmic domain | ||||||||||||
 Sample Sample |
| ||||||||||||
| Function / homology | Mce/MlaD / MlaD protein / intermembrane lipid transfer / outer membrane-bounded periplasmic space / identical protein binding /  plasma membrane / Intermembrane transport protein YebT plasma membrane / Intermembrane transport protein YebT Function and homology information Function and homology information | ||||||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 25.0 Å negative staining / Resolution: 25.0 Å | ||||||||||||
 Authors Authors | Bhabha G / Ekiert DC | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: Architectures of Lipid Transport Systems for the Bacterial Outer Membrane. Authors: Damian C Ekiert / Gira Bhabha / Georgia L Isom / Garrett Greenan / Sergey Ovchinnikov / Ian R Henderson / Jeffery S Cox / Ronald D Vale /   Abstract: How phospholipids are trafficked between the bacterial inner and outer membranes through the hydrophilic space of the periplasm is not known. We report that members of the mammalian cell entry (MCE) ...How phospholipids are trafficked between the bacterial inner and outer membranes through the hydrophilic space of the periplasm is not known. We report that members of the mammalian cell entry (MCE) protein family form hexameric assemblies with a central channel capable of mediating lipid transport. The E. coli MCE protein, MlaD, forms a ring associated with an ABC transporter complex in the inner membrane. A soluble lipid-binding protein, MlaC, ferries lipids between MlaD and an outer membrane protein complex. In contrast, EM structures of two other E. coli MCE proteins show that YebT forms an elongated tube consisting of seven stacked MCE rings, and PqiB adopts a syringe-like architecture. Both YebT and PqiB create channels of sufficient length to span the periplasmic space. This work reveals diverse architectures of highly conserved protein-based channels implicated in the transport of lipids between the membranes of bacteria and some eukaryotic organelles. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8611.map.gz emd_8611.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8611-v30.xml emd-8611-v30.xml emd-8611.xml emd-8611.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8611.png emd_8611.png | 57.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8611 http://ftp.pdbj.org/pub/emdb/structures/EMD-8611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8611 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8611 | HTTPS FTP |
-Related structure data
| Related structure data |  8608C  8610C 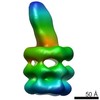 8612C  5uvnC  5uw2C  5uw8C  5uwaC  5uwbC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8611.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8611.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E. coli MCE protein PqiB, periplasmic domain | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : YebT
| Entire | Name: YebT |
|---|---|
| Components |
|
-Supramolecule #1: YebT
| Supramolecule | Name: YebT / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: YebT
| Macromolecule | Name: YebT / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MHMSQETPAS TTEAQIKNKR RISPFWLLP FIALMIASWL I WDSYQDRG NTVTIDFMSA DG IVPGRTP VRYQGVEVGT VQD ISLSDD LRKIEVKVSI KSDM KDALR EETQFWLVTP KASLA GVSG LDALVGGNYI GMMPGK GKE QDHFVALDTQ PKYRLDN GD ...String: MHMSQETPAS TTEAQIKNKR RISPFWLLP FIALMIASWL I WDSYQDRG NTVTIDFMSA DG IVPGRTP VRYQGVEVGT VQD ISLSDD LRKIEVKVSI KSDM KDALR EETQFWLVTP KASLA GVSG LDALVGGNYI GMMPGK GKE QDHFVALDTQ PKYRLDN GD LMIHLQAPDL GSLNSGSL V YFRKIPVGKV YDYAINPNK QGVVIDVLIE RRFTDLVKKG SRFWNVSGV DANVSISGAK V KLESLAAL VNGAIAFDSP EE SKPAEAE DTFGLYEDLA HSQ RGVIIK LELPSGAGLT ADST PLMYQ GLEVGQLTKL DLNPG GKVT GEMTVDPSVV TLLREN TRI ELRNPKLSLS DANLSAL LT GKTFELVPGD GEPRKEFV V VPGEKALLHE PDVLTLTLT APESYGIDAG QPLILHGVQV GQVIDRKLT SKGVTFTVAI E PQHRELVK GDSKFVVNSR VD VKVGLDG VEFLGASASE WIN GGIRIL PGDKGEMKAS YPLY ANLEK ALENSLSDLP TTTVS LSAE TLPDVQAGSV VLYRKF EVG EVITVRPRAN AFDIDLH IK PEYRNLLTSN SVFWAEGG A KVQLNGSGLT VQASPLSRA LKGAISFDNL SGASASQRKG DKRILYASE TAARAVGGQI T LHAFDAGK LAVGMPIRYL GI DIGQIQT LDLITARNEV QAK AVLYPE YVQTFARGGT RFSV VTPQI SAAGVEHLDT ILQPY INVE PGRGNPRRDF ELQEAT ITD SRYLDGLSII VEAPEAG SL GIGTPVLFRG LEVGTVTG M TLGTLSDRVM IAMRISKRY QHLVRNNSVF WLASGYSLDF GLTGGVVKT GTFNQFIRGG I AFATPPGT PLAPKAQEGK HF LLQESEP KEWREWGTAL PKH QHQHHH HHH |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 10.0 e/Å2 |
- Image processing
Image processing
| Initial angle assignment | Type: RANDOM ASSIGNMENT / Software - Name: RELION (ver. 1.4) |
|---|---|
| Final 3D classification | Software - Name: RELION (ver. 1.4) |
| Final angle assignment | Type: OTHER / Software - Name: RELION (ver. 1.4) |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 4787 |
 Movie
Movie Controller
Controller










