[English] 日本語
 Yorodumi
Yorodumi- EMDB-8576: Structure of the Plasmodium falciparum 80S ribosome bound to the ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8576 | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Plasmodium falciparum 80S ribosome bound to the antimalarial drug mefloquine | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationTranslesion synthesis by REV1 / Translesion Synthesis by POLH / : / Translesion synthesis by POLK / Translesion synthesis by POLI / Josephin domain DUBs / : / Metalloprotease DUBs / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / ER Quality Control Compartment (ERQC) ...Translesion synthesis by REV1 / Translesion Synthesis by POLH / : / Translesion synthesis by POLK / Translesion synthesis by POLI / Josephin domain DUBs / : / Metalloprotease DUBs / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / ER Quality Control Compartment (ERQC) / Iron uptake and transport / L13a-mediated translational silencing of Ceruloplasmin expression / SRP-dependent cotranslational protein targeting to membrane / Major pathway of rRNA processing in the nucleolus and cytosol / Formation of a pool of free 40S subunits / Formation of the ternary complex, and subsequently, the 43S complex / Ribosomal scanning and start codon recognition / GTP hydrolysis and joining of the 60S ribosomal subunit / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Aggrephagy / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / : / Orc1 removal from chromatin / CDK-mediated phosphorylation and removal of Cdc6 / KEAP1-NFE2L2 pathway / UCH proteinases / Ub-specific processing proteases /  Neddylation / Antigen processing: Ubiquitination & Proteasome degradation / protein-RNA complex assembly / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of LSU-rRNA / Neddylation / Antigen processing: Ubiquitination & Proteasome degradation / protein-RNA complex assembly / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of LSU-rRNA /  ribosomal large subunit biogenesis / modification-dependent protein catabolic process / ribosomal large subunit biogenesis / modification-dependent protein catabolic process /  protein tag activity / protein tag activity /  ribosomal large subunit assembly / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / large ribosomal subunit / ribosomal large subunit assembly / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / large ribosomal subunit /  ribosome biogenesis / cytoplasmic translation / ribosome biogenesis / cytoplasmic translation /  5S rRNA binding / cytosolic large ribosomal subunit / ubiquitin-dependent protein catabolic process / negative regulation of translation / 5S rRNA binding / cytosolic large ribosomal subunit / ubiquitin-dependent protein catabolic process / negative regulation of translation /  rRNA binding / protein ubiquitination / rRNA binding / protein ubiquitination /  ribosome / structural constituent of ribosome / ribosome / structural constituent of ribosome /  ribonucleoprotein complex / ribonucleoprotein complex /  translation / translation /  mRNA binding / mRNA binding /  ubiquitin protein ligase binding / ubiquitin protein ligase binding /  RNA binding / RNA binding /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | |||||||||||||||||||||||||||
| Biological species |   Plasmodium falciparum 3D7 (eukaryote) Plasmodium falciparum 3D7 (eukaryote) | |||||||||||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||||||||||||||||||||
 Authors Authors | Wong W / Bai X-C / Brown A / Scheres S / Baum J | |||||||||||||||||||||||||||
| Funding support |  Australia, Australia,  United Kingdom, 8 items United Kingdom, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2017 Journal: Nat Microbiol / Year: 2017Title: Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Authors: Wilson Wong / Xiao-Chen Bai / Brad E Sleebs / Tony Triglia / Alan Brown / Jennifer K Thompson / Katherine E Jackson / Eric Hanssen / Danushka S Marapana / Israel S Fernandez / Stuart A Ralph ...Authors: Wilson Wong / Xiao-Chen Bai / Brad E Sleebs / Tony Triglia / Alan Brown / Jennifer K Thompson / Katherine E Jackson / Eric Hanssen / Danushka S Marapana / Israel S Fernandez / Stuart A Ralph / Alan F Cowman / Sjors H W Scheres / Jake Baum /   Abstract: Malaria control is heavily dependent on chemotherapeutic agents for disease prevention and drug treatment. Defining the mechanism of action for licensed drugs, for which no target is characterized, ...Malaria control is heavily dependent on chemotherapeutic agents for disease prevention and drug treatment. Defining the mechanism of action for licensed drugs, for which no target is characterized, is critical to the development of their second-generation derivatives to improve drug potency towards inhibition of their molecular targets. Mefloquine is a widely used antimalarial without a known mode of action. Here, we demonstrate that mefloquine is a protein synthesis inhibitor. We solved a 3.2 Å cryo-electron microscopy structure of the Plasmodium falciparum 80S ribosome with the (+)-mefloquine enantiomer bound to the ribosome GTPase-associated centre. Mutagenesis of mefloquine-binding residues generates parasites with increased resistance, confirming the parasite-killing mechanism. Furthermore, structure-guided derivatives with an altered piperidine group, predicted to improve binding, show enhanced parasiticidal effect. These data reveal one possible mode of action for mefloquine and demonstrate the vast potential of cryo-electron microscopy to guide the development of mefloquine derivatives to inhibit parasite protein synthesis. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8576.map.gz emd_8576.map.gz | 319.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8576-v30.xml emd-8576-v30.xml emd-8576.xml emd-8576.xml | 64.3 KB 64.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8576.png emd_8576.png | 86.6 KB | ||
| Others |  emd_8576_half_map_1.map.gz emd_8576_half_map_1.map.gz emd_8576_half_map_2.map.gz emd_8576_half_map_2.map.gz | 140.5 MB 140.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8576 http://ftp.pdbj.org/pub/emdb/structures/EMD-8576 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8576 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8576 | HTTPS FTP |
-Related structure data
| Related structure data |  5umdMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8576.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8576.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
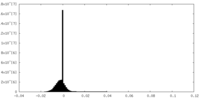
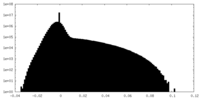
-Half map: #1
| File | emd_8576_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
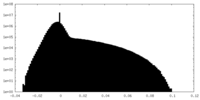
-Half map: #2
| File | emd_8576_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Plasmodium falciparum 80S ribosome bound to mefloquine
+Supramolecule #1: Plasmodium falciparum 80S ribosome bound to mefloquine
+Macromolecule #1: 28S ribosomal RNA
+Macromolecule #2: 5S ribosomal RNA
+Macromolecule #3: 5.8S ribosomal RNA
+Macromolecule #4: 60S ribosomal protein L2
+Macromolecule #5: 60S ribosomal protein L3
+Macromolecule #6: 60S ribosomal protein L4
+Macromolecule #7: 60S ribosomal protein L11a, putative
+Macromolecule #8: 60S ribosomal protein L6, putative
+Macromolecule #9: 60S ribosomal protein L6-2, putative
+Macromolecule #10: 60S ribosomal protein L7-3, putative
+Macromolecule #11: 60S ribosomal protein L13, putative
+Macromolecule #12: 60S ribosomal protein L13
+Macromolecule #13: 60S ribosomal protein L23, putative
+Macromolecule #14: 60S ribosomal protein L14, putative
+Macromolecule #15: 60S ribosomal protein L27a, putative
+Macromolecule #16: Ribosomal protein L15
+Macromolecule #17: 60S ribosomal protein L10, putative
+Macromolecule #18: 60S ribosomal protein L5, putative
+Macromolecule #19: 60S ribosomal protein L18-2, putative
+Macromolecule #20: 60S ribosomal protein L19
+Macromolecule #21: 60S ribosomal protein L18a
+Macromolecule #22: 60S ribosomal protein L21
+Macromolecule #23: 60S ribosomal protein L17, putative
+Macromolecule #24: 60S ribosomal protein L22, putative
+Macromolecule #25: 60S ribosomal protein L23
+Macromolecule #26: 60S ribosomal protein L26, putative
+Macromolecule #27: 60S ribosomal protein L24, putative
+Macromolecule #28: 60S ribosomal protein L27
+Macromolecule #29: 60S ribosomal protein L28
+Macromolecule #30: 60S ribosomal protein L35, putative
+Macromolecule #31: 60S ribosomal protein L29, putative
+Macromolecule #32: 60S ribosomal protein L7, putative
+Macromolecule #33: 60S ribosomal protein L30e, putative
+Macromolecule #34: 60S ribosomal protein L31
+Macromolecule #35: 60S ribosomal protein L32
+Macromolecule #36: 60S ribosomal protein L35Ae, putative
+Macromolecule #37: 60S ribosomal protein L34
+Macromolecule #38: 60S ribosomal protein L36
+Macromolecule #39: Ribosomal protein L37
+Macromolecule #40: 60S ribosomal protein L38
+Macromolecule #41: 60S ribosomal protein L39
+Macromolecule #42: Ubiquitin-60S ribosomal protein L40
+Macromolecule #43: 60S ribosomal protein L41
+Macromolecule #44: 60S ribosomal protein L37a
+Macromolecule #45: 60S ribosomal protein L44
+Macromolecule #46: Mefloquine
+Macromolecule #47: MAGNESIUM ION
+Macromolecule #48: ZINC ION
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: 20 mM Hepes pH 7.4, 40 mM KAc, 10 mM NH4Ac, 10 mM Mg(Ac)2 and 5 mM 2-mecaptoethanol |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.02 nm / Pretreatment - Type: GLOW DISCHARGE / Details: Blot 2.5s before plunging |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 90 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.8 µm / Calibrated defocus min: 0.8 µm / Calibrated magnification: 104748 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm / Nominal magnification: 78000 Bright-field microscopy / Cs: 2.0 mm / Nominal magnification: 78000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 80.0 K / Max: 90.0 K |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Number grids imaged: 1 / Average electron dose: 20.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: CTFFIND (ver. 3) |
|---|---|
| Startup model | Type of model: EMDB MAP EMDB ID: |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 1.3) |
| Final 3D classification | Number classes: 4 / Software - Name: RELION (ver. 1.3) |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 1.3) |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.3) / Number images used: 43184 |
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Overall B value: 129.3 |
|---|---|
| Output model |  PDB-5umd: |
 Movie
Movie Controller
Controller




















 Z
Z Y
Y X
X