+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6803 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
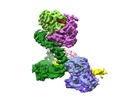
| Title | Cryo-EM structure of Human DNA-PK Holoenzyme | |||||||||||||||
 Map data Map data | None | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Cryo-EM structure / DNA-PK / DNAPKcs /  activation / activation /  NHEJ / NHEJ /  DNA BINDING PROTEIN DNA BINDING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationKu70:Ku80 complex / negative regulation of t-circle formation / positive regulation of platelet formation /  DNA end binding / T cell receptor V(D)J recombination / pro-B cell differentiation / small-subunit processome assembly / positive regulation of lymphocyte differentiation / DNA-dependent protein kinase activity / histone H2AXS139 kinase activity ...Ku70:Ku80 complex / negative regulation of t-circle formation / positive regulation of platelet formation / DNA end binding / T cell receptor V(D)J recombination / pro-B cell differentiation / small-subunit processome assembly / positive regulation of lymphocyte differentiation / DNA-dependent protein kinase activity / histone H2AXS139 kinase activity ...Ku70:Ku80 complex / negative regulation of t-circle formation / positive regulation of platelet formation /  DNA end binding / T cell receptor V(D)J recombination / pro-B cell differentiation / small-subunit processome assembly / positive regulation of lymphocyte differentiation / DNA-dependent protein kinase activity / histone H2AXS139 kinase activity / DNA-dependent protein kinase complex / immature B cell differentiation / DNA-dependent protein kinase-DNA ligase 4 complex / cellular response to X-ray / immunoglobulin V(D)J recombination / DNA end binding / T cell receptor V(D)J recombination / pro-B cell differentiation / small-subunit processome assembly / positive regulation of lymphocyte differentiation / DNA-dependent protein kinase activity / histone H2AXS139 kinase activity / DNA-dependent protein kinase complex / immature B cell differentiation / DNA-dependent protein kinase-DNA ligase 4 complex / cellular response to X-ray / immunoglobulin V(D)J recombination /  nonhomologous end joining complex / nonhomologous end joining complex /  DNA ligation / nuclear telomere cap complex / regulation of smooth muscle cell proliferation / double-strand break repair via alternative nonhomologous end joining / Cytosolic sensors of pathogen-associated DNA / double-strand break repair via classical nonhomologous end joining / regulation of epithelial cell proliferation / IRF3-mediated induction of type I IFN / telomere capping / DNA ligation / nuclear telomere cap complex / regulation of smooth muscle cell proliferation / double-strand break repair via alternative nonhomologous end joining / Cytosolic sensors of pathogen-associated DNA / double-strand break repair via classical nonhomologous end joining / regulation of epithelial cell proliferation / IRF3-mediated induction of type I IFN / telomere capping /  recombinational repair / recombinational repair /  regulation of telomere maintenance / U3 snoRNA binding / regulation of hematopoietic stem cell differentiation / positive regulation of neurogenesis / protein localization to chromosome, telomeric region / cellular response to fatty acid / hematopoietic stem cell proliferation / cellular hyperosmotic salinity response / maturation of 5.8S rRNA / T cell lineage commitment / negative regulation of cGAS/STING signaling pathway / telomeric DNA binding / positive regulation of catalytic activity / B cell lineage commitment / 2-LTR circle formation / positive regulation of double-strand break repair via nonhomologous end joining / site of DNA damage / regulation of telomere maintenance / U3 snoRNA binding / regulation of hematopoietic stem cell differentiation / positive regulation of neurogenesis / protein localization to chromosome, telomeric region / cellular response to fatty acid / hematopoietic stem cell proliferation / cellular hyperosmotic salinity response / maturation of 5.8S rRNA / T cell lineage commitment / negative regulation of cGAS/STING signaling pathway / telomeric DNA binding / positive regulation of catalytic activity / B cell lineage commitment / 2-LTR circle formation / positive regulation of double-strand break repair via nonhomologous end joining / site of DNA damage /  Lyases; Carbon-oxygen lyases; Other carbon-oxygen lyases / Lyases; Carbon-oxygen lyases; Other carbon-oxygen lyases /  enzyme activator activity / 5'-deoxyribose-5-phosphate lyase activity / hematopoietic stem cell differentiation / positive regulation of protein kinase activity / ectopic germ cell programmed cell death / ATP-dependent activity, acting on DNA / enzyme activator activity / 5'-deoxyribose-5-phosphate lyase activity / hematopoietic stem cell differentiation / positive regulation of protein kinase activity / ectopic germ cell programmed cell death / ATP-dependent activity, acting on DNA /  somitogenesis / positive regulation of telomerase activity / mitotic G1 DNA damage checkpoint signaling / positive regulation of telomere maintenance via telomerase / somitogenesis / positive regulation of telomerase activity / mitotic G1 DNA damage checkpoint signaling / positive regulation of telomere maintenance via telomerase /  DNA helicase activity / activation of innate immune response / DNA helicase activity / activation of innate immune response /  telomere maintenance / cellular response to leukemia inhibitory factor / telomere maintenance / cellular response to leukemia inhibitory factor /  neurogenesis / small-subunit processome / neurogenesis / small-subunit processome /  cyclin binding / positive regulation of translation / negative regulation of protein phosphorylation / positive regulation of erythrocyte differentiation / cyclin binding / positive regulation of translation / negative regulation of protein phosphorylation / positive regulation of erythrocyte differentiation /  protein-DNA complex / response to gamma radiation / Nonhomologous End-Joining (NHEJ) / protein-DNA complex / response to gamma radiation / Nonhomologous End-Joining (NHEJ) /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement /  brain development / peptidyl-threonine phosphorylation / protein destabilization / protein modification process / cellular response to gamma radiation / brain development / peptidyl-threonine phosphorylation / protein destabilization / protein modification process / cellular response to gamma radiation /  regulation of circadian rhythm / double-strand break repair via nonhomologous end joining / cellular response to insulin stimulus / rhythmic process / double-strand break repair / intrinsic apoptotic signaling pathway in response to DNA damage / E3 ubiquitin ligases ubiquitinate target proteins / T cell differentiation in thymus / regulation of circadian rhythm / double-strand break repair via nonhomologous end joining / cellular response to insulin stimulus / rhythmic process / double-strand break repair / intrinsic apoptotic signaling pathway in response to DNA damage / E3 ubiquitin ligases ubiquitinate target proteins / T cell differentiation in thymus /  heart development / heart development /  double-stranded DNA binding / double-stranded DNA binding /  scaffold protein binding / peptidyl-serine phosphorylation / secretory granule lumen / DNA recombination / ficolin-1-rich granule lumen / scaffold protein binding / peptidyl-serine phosphorylation / secretory granule lumen / DNA recombination / ficolin-1-rich granule lumen /  transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / damaged DNA binding / transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / damaged DNA binding /  chromosome, telomeric region / transcription cis-regulatory region binding / chromosome, telomeric region / transcription cis-regulatory region binding /  non-specific serine/threonine protein kinase / non-specific serine/threonine protein kinase /  protein kinase activity / response to xenobiotic stimulus / protein kinase activity / response to xenobiotic stimulus /  ribonucleoprotein complex / positive regulation of apoptotic process / protein domain specific binding / ribonucleoprotein complex / positive regulation of apoptotic process / protein domain specific binding /  protein phosphorylation protein phosphorylationSimilarity search - Function | |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
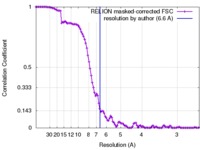
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 6.6 Å cryo EM / Resolution: 6.6 Å | |||||||||||||||
 Authors Authors | Yin X / Liu M | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2017 Journal: Cell Res / Year: 2017Title: Cryo-EM structure of human DNA-PK holoenzyme. Authors: Xiaotong Yin / Mengjie Liu / Yuan Tian / Jiawei Wang / Yanhui Xu /  Abstract: DNA-dependent protein kinase (DNA-PK) is a serine/threonine protein kinase complex composed of a catalytic subunit (DNA-PKcs) and KU70/80 heterodimer bound to DNA. DNA-PK holoenzyme plays a critical ...DNA-dependent protein kinase (DNA-PK) is a serine/threonine protein kinase complex composed of a catalytic subunit (DNA-PKcs) and KU70/80 heterodimer bound to DNA. DNA-PK holoenzyme plays a critical role in non-homologous end joining (NHEJ), the major DNA repair pathway. Here, we determined cryo-electron microscopy structure of human DNA-PK holoenzyme at 6.6 Å resolution. In the complex structure, DNA-PKcs, KU70, KU80 and DNA duplex form a 650-kDa heterotetramer with 1:1:1:1 stoichiometry. The N-terminal α-solenoid (∼2 800 residues) of DNA-PKcs adopts a double-ring fold and connects the catalytic core domain of DNA-PKcs and KU70/80-DNA. DNA-PKcs and KU70/80 together form a DNA-binding tunnel, which cradles ∼30-bp DNA and prevents sliding inward of DNA-PKcs along with DNA duplex, suggesting a mechanism by which the broken DNA end is protected from unnecessary processing. Structural and biochemical analyses indicate that KU70/80 and DNA coordinately induce conformational changes of DNA-PKcs and allosterically stimulate its kinase activity. We propose a model for activation of DNA-PKcs in which allosteric signals are generated upon DNA-PK holoenzyme formation and transmitted to the kinase domain through N-terminal HEAT repeats and FAT domain of DNA-PKcs. Our studies suggest a mechanism for recognition and protection of broken DNA ends and provide a structural basis for understanding the activation of DNA-PKcs and DNA-PK-mediated NHEJ pathway. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6803.map.gz emd_6803.map.gz | 6.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6803-v30.xml emd-6803-v30.xml emd-6803.xml emd-6803.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6803_fsc.xml emd_6803_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_6803.png emd_6803.png | 116.9 KB | ||
| Filedesc metadata |  emd-6803.cif.gz emd-6803.cif.gz | 9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6803 http://ftp.pdbj.org/pub/emdb/structures/EMD-6803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6803 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6803 | HTTPS FTP |
-Related structure data
| Related structure data |  5y3rMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6803.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6803.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : PRKDC-Helix
| Entire | Name: PRKDC-Helix |
|---|---|
| Components |
|
-Supramolecule #1: PRKDC-Helix
| Supramolecule | Name: PRKDC-Helix / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: X-ray repair cross-complementing protein 6
| Macromolecule | Name: X-ray repair cross-complementing protein 6 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number:  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.619352 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GRDSLIFLVD ASKAMFESQS EDELTPFDMS IQCIQSVYIS KIISSDRDLL AVVFYGTEKD KNSVNFKNIY VLQELDNPGA KRILELDQF KGQQGQKRFQ DMMGHGSDYS LSEVLWVCAN LFSDVQFKMS HKRIMLFTNE DNPHGNDSAK ASRARTKAGD L RDTGIFLD ...String: GRDSLIFLVD ASKAMFESQS EDELTPFDMS IQCIQSVYIS KIISSDRDLL AVVFYGTEKD KNSVNFKNIY VLQELDNPGA KRILELDQF KGQQGQKRFQ DMMGHGSDYS LSEVLWVCAN LFSDVQFKMS HKRIMLFTNE DNPHGNDSAK ASRARTKAGD L RDTGIFLD LMHLKKPGGF DISLFYRDII SIAEDEDLRV HFEESSKLED LLRKVRAKET RKRALSRLKL KLNKDIVISV GI YNLVQKA LKPPPIKLYR ETNEPVKTKT RTFNTSTGGL LLPSDTKRSQ IYGSRQIILE KEETEELKRF DDPGLMLMGF KPL VLLKKH HYLRPSLFVY PEESLVIGSS TLFSALLIKC LEKEVAALCR YTPRRNIPPY FVALVPQEEE LDDQKIQVTP PGFQ LVFLP FADDKRKMPF TEKIMATPEQ VGKMKAIVEK LRFTYRSDSF ENPVLQQHFR NLEALALDLM EPEQAVDLTL PKVEA MNKR LGSLVDEFKE LVYPPDY UniProtKB: X-ray repair cross-complementing protein 6 |
-Macromolecule #2: X-ray repair cross-complementing protein 5
| Macromolecule | Name: X-ray repair cross-complementing protein 5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number:  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.972969 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NKAAVVLCMD VGFTMSNSIP GIESPFEQAK KVITMFVQRQ VFAENKDEIA LVLFGTDGTD NPLSGGDQYQ NITVHRHLML PDFDLLEDI ESKIQPGSQQ ADFLDALIVS MDVIQHETIG KKFEKRHIEI FTDLSSRFSK SQLDIIIHSL KKCDISLQFF L PFSLGKED ...String: NKAAVVLCMD VGFTMSNSIP GIESPFEQAK KVITMFVQRQ VFAENKDEIA LVLFGTDGTD NPLSGGDQYQ NITVHRHLML PDFDLLEDI ESKIQPGSQQ ADFLDALIVS MDVIQHETIG KKFEKRHIEI FTDLSSRFSK SQLDIIIHSL KKCDISLQFF L PFSLGKED GSGDRGDGPF RLGGHGPSFP LKGITEQQKE GLEIVKMVMI SLEGEDGLDE IYSFSESLRK LCVFKKIERH SI HWPCRLT IGSNLSIRIA AYKSILQERV KKTWTVVDAK TLKKEDIQKE TVYCLNDDDE TEVLKEDIIQ GFRYGSDIVP FSK VDEEQM KYKSEGKCFS VLGFCKSSQV QRRFFMGNQV LKVFAARDDE AAAVALSSLI HALDDLDMVA IVRYAYDKRA NPQV GVAFP HIKHNYECLV YVQLPFMEDL RQYMFSSLKN SKKYAPTEAQ LNAVDALIDS MSLAKKDEKT DTLEDLFPTT KIPNP RFQR LFQCLLHRAL HPREPLPPIQ QHIWNMLNPP AEVTTKSQIP LSKIKTLFPL IE UniProtKB: X-ray repair cross-complementing protein 5 |
-Macromolecule #3: PRKDC-Helix
| Macromolecule | Name: PRKDC-Helix / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.084182 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AAAAAAAAAA AAAAA |
-Macromolecule #6: DNA-dependent protein kinase catalytic subunit
| Macromolecule | Name: DNA-dependent protein kinase catalytic subunit / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number:  non-specific serine/threonine protein kinase non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 468.885344 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: CSLLRLQETL SAADRCGAAL AGHQLIRGLG QECVLSSSPA VLALQTSLVF SRDFGLLVFV RKSLNSIEFR ECREEILKFL CIFLEKMGQ KIAPYSVEIK NTCTSVYTKD RAAKCKIPAL DLLIKLLQTF RSSRLMDEFK IGELFSKFYG ELALKKKIPD T VLEKVYEL ...String: CSLLRLQETL SAADRCGAAL AGHQLIRGLG QECVLSSSPA VLALQTSLVF SRDFGLLVFV RKSLNSIEFR ECREEILKFL CIFLEKMGQ KIAPYSVEIK NTCTSVYTKD RAAKCKIPAL DLLIKLLQTF RSSRLMDEFK IGELFSKFYG ELALKKKIPD T VLEKVYEL LGLLGEVHPS EMINNAENLF RAFLGELKTQ MTSAVREPKL PVLAGCLKGL SSLLCNFTKS MEEDPQTSRE IF NFVLKAI RPQIDLKRYA VPSAGLRLFA LHASQFSTCL LDNYVSLFEV LLKWCAHTNV ELKKAALSAL ESFLKQVSNM VAK NAEMHK NKLQYFMEQF YGIIRNVDSN NKELSIAIRG YGLFAGPCKV INAKDVDFMY VELIQRCKQM FLTQTDTGDD RVYQ MPSFL QSVASVLLYL DTVPEVYTPV LEHLVVMQID SFPQYSPKMQ LVCCRAIVKV FLALAAKGPV LRNCISTVVH QGLIR ICSK PVVLPKGPES ESEDHRASGE VRTGKWKVPT YKDYVDLFRH LLSSDQMMDS ILADEAFFSV NSSSESLNHL LYDEFV KSV LKIVEKLDLT LEIQTVGEQE NGDEAPGVWM IPTSDPAANL HPAKPKDFSA FINLVEFCRE ILPEKQAEFF EPWVYSF SY ELILQSTRLP LISGFYKLLS ITVRNAKKIK YFEGVSPKSL KHSPEDPEKY SCFALFVKFG KEVAVKMKQY KDELLASC L TFLLSLPHNI IELDVRAYVP ALQMAFKLGL SYTPLAEVGL NALEEWSIYI DRHVMQPYYK DILPCLDGYL KTSALSDET KNNWEVSALS RAAQKGFNKV VLKHLKKTKN LSSNEAISLE EIRIRVVQML GSLGGQINKN LLTVTSSDEM MKSYVAWDRE KRLSFAVPF REMKPVIFLD VFLPRVTELA LTASDRQTKV AACELLHSMV MFMLGKATQM PEGGQGAPPM YQLYKRTFPV L LRLACDVD QVTRQLYEPL VMQLIHWFTN NKKFESQDTV ALLEAILDGI VDPVDSTLRD FCGRCIREFL KWSIKQITPQ QQ EKSPVNT KSLFKRLYSL ALHPNAFKRL GASLAFNNIY REFREEESLV EQFVFEALVI YMESLALAHA DEKSLGTIQQ CCD AIDHLC RIIEKKHVSL NKAKKRRLPR GFPPSASLCL LDLVKWLLAH CGRPQTECRH KSIELFYKFV PLLPGNRSPN LWLK DVLKE EGVSFLINTF EGGGCGQPSG ILAQPTLLYL RGPFSLQATL CWLDLLLAAL ECYNTFIGER TVGALQVLGT EAQSS LLKA VAFFLESIAM HDIIAAEKCF GTGAAGNRTS PQEGERYNYS KCTVVVRIME FTTTLLNTSP EGWKLLKKDL CNTHLM RVL VQTLCEPASI GFNIGDVQVM AHLPDVCVNL MKALKMSPYK DILETHLREK ITAQSIEELC AVNLYGPDAQ VDRSRLA AV VSACKQLHRA GLLHNILPSQ STDLHHSVGT ELLSLVYKGI APGDERQCLP SLDLSCKQLA SGLLELAFAF GGLCERLV S LLLNPAVLST ASLGSSQGSV IHFSHGEYFY SLFSETINTE LLKNLDLAVL ELMQSSVDNT KMVSAVLNGM LDQSFRERA NQKHQGLKLA TTILQHWKKC DSWWAKDSPL ETKMAVLALL AKILQIDSSV SFNTSHGSFP EVFTTYISLL ADTKLDLHLK GQAVTLLPF FTSLTGGSLE ELRRVLEQLI VAHFPMQSRE FPPGTPRFNN YVDCMKKFLD ALELSQSPML LELMTEVLCR E QQHVMEEL FQSSFRRIAR RGSCVTQVGL LESVYEMFRK DDPRLSFTRQ SFVDRSLLTL LWHCSLDALR EFFSTIVVDA ID VLKSRFT KLNESTFDTQ ITKKMGYYKI LDVMYSRLPK DDVHAKESKI NQVFHGSCIT EGNELTKTLI KLCYDAFTEN MAG ENQLLE RRRLYHCAAY NCAISVICCV FNELKFYQGF LFSEKPEKNL LIFENLIDLK RRYNFPVEVE VPMERKKKYI EIRK EAREA ANGDSDGPSY MSSLSYLADS TLSEEMSQFD FSTGVQSYSY SSQDPRPATG RFRRREQRDP TVHDDVLELE MDELN RHEC MAPLTALVKH MHRSLGPPQG EEDSVPRDLP SWMKFLHGKL GNPIVPLNIR LFLAKLVINT EEVFRPYAKH WLSPLL QLA ASENNGGEGI HYMVVEIVAT ILSWTGLATP TGVPKDEVLA NRLLNFLMKH VFHPKRAVFR HNLEIIKTLV ECWKDCL SI PYRLIFEKFS GKDPNSKDNS VGIQLLGIVM ANDLPPYDPQ CGIQSSEYFQ ALVNNMSFVR YKEVYAAAAE VLGLILRY V MERKNILEES LCELVAKQLK QHQNTMEDKF IVCLNKVTKS FPPLADRFMN AVFFLLPKFH GVLKTLCLEV VLCRVEGMT ELYFQLKSKD FVQVMRHRDD ERQKVCLDII YKMMPKLKPV ELRELLNPVV EFVSHPSTTC REQMYNILMW IHDNYRDPES ETDNDSQEI FKLAKDVLIQ GLIDENPGLQ LIIRNFWSHE TRLPSNTLDR LLALNSLYSP KIEVHFLSLA TNFLLEMTSM S PDYPNPMF EHPLSECEFQ EYTIDSDWRF RSTVLTPMFV ETQASQGTLQ TRTQEGSLSA RWPVAGQIRA TQQQHDFTLT QT ADGRSSF DWLTGSSTDP LVDHTSPSSD SLLFAHKRSE RLQRAPLKSV GPDFGKKRLG LPGDEVDNKV KGAAGRTDLL RLR RRFMRD QEKLSLMYAR KGVAEQKREK EIKSELKMKQ DAQVVLYRSY RHGDLPDIQI KHSSLITPLQ AVAQRDPIIA KQLF SSLFS GILKEMDKFK TLSEKNNITQ KLLQDFNRFL NTTFSFFPPF VSCIQDISCQ HAALLSLDPA AVSAGCLASL QQPVG IRLL EEALLRLLPA ELPAKRVRGK ARLPPDVLRW VELAKLYRSI GEYDVLRGIF TSEIGTKQIT QSALLAEARS DYSEAA KQY DEALNKQDWV DGEPTEAEKD FWELASLDCY NHLAEWKSLE YCSTASIDSE NPPDLNKIWS EPFYQETYLP YMIRSKL KL LLQGEADQSL LTFIDKAMHG ELQKAILELH YSQELSLLYL LQDDVDRAKY YIQNGIQSFM QNYSSIDVLL HQSRLTKL Q SVQALTEIQE FISFISKQGN LSSQVPLKRL LNTWTNRYPD AKMDPMNIWD DIITNRCFFL SKIEEKLTPL PEDNSMNVD QDGDPSDRME VQEQEEDISS LIRSCKFSMK MKMIDSARKQ NNFSLAMKLL KELHKESKTR DDWLVSWVQS YCRLSHCRSR SQGCSEQVL TVLKTVSLLD ENNVSSYLSK NILAFRDQNI LLGTTYRIIA NALSSEPACL AEIEEDKARR ILELSGSSSE D SEKVIAGL YQRAFQHLSE AVQAAEEEAQ PPSWSCGPAA GVIDAYMTLA DFCDQQLRKE EENASVIDSA ELQAYPALVV EK MLKALKL NSNEARLKFP RLLQIIERYP EETLSLMTKE ISSVPCWQFI SWISHMVALL DKDQAVAVQH SVEEITDNYP QAI VYPFII SSESYSFKDT STGHKNKEFV ARIKSKLDQG GVIQDFINAL DQLSNPELLF KDWSNDVRAE LAKTPVNKKN IEKM YERMY AALGDPKAPG LGAFRRKFIQ TFGKEFDKHF GKGGSKLLRM KLSDFNDITN MLLLKMNKDS KPPGNLKECS PWMSD FKVE FLRNELEIPG QYDGRGKPLP EYHVRIAGFD ERVTVMASLR RPKRIIIRGH DEREHPFLVK GGEDLRQDQR VEQLFQ VMN GILAQDSACS QRALQLRTYS VVPMTSRLGL IEWLENTVTL KDLLLNTMSQ EEKAAYLSDP RAPPCEYKDW LTKMSGK HD VGAYMLMYKG ANRTETVTSF RKRESKVPAD LLKRAFVRMS TSPEAFLALR SHFASSHALI CISHWILGIG DRHLNNFM V AMETGGVIGI DFGHAFGSAT QFLPVPELMP FRLTRQFINL MLPMKETGLM YSIMVHALRA FRSDPGLLTN TMDVFVKEP SFDWKNFEQK MLKKGGSWIQ EINVAEKNWY PRQKICYAKR KLAGANPAVI TCDELLLGHE KAPAFRDYVA VARGSKDHNI RAQEPESGL SEETQVKCLM DQATDPNILG RTWEGWEPWM UniProtKB: DNA-dependent protein kinase catalytic subunit |
-Macromolecule #4: DNA (34-MER)
| Macromolecule | Name: DNA (34-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.502785 KDa |
| Sequence | String: (DT)(DA)(DA)(DA)(DA)(DA)(DC)(DT)(DA)(DT) (DT)(DA)(DT)(DT)(DA)(DT)(DG)(DG)(DT)(DA) (DT)(DT)(DA)(DT)(DG)(DG)(DC)(DC)(DT) (DT)(DG)(DG)(DG)(DC) |
-Macromolecule #5: DNA (36-MER)
| Macromolecule | Name: DNA (36-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.042164 KDa |
| Sequence | String: (DC)(DA)(DG)(DC)(DT)(DA)(DA)(DT)(DG)(DG) (DC)(DC)(DA)(DT)(DA)(DA)(DT)(DA)(DC)(DC) (DA)(DT)(DA)(DA)(DT)(DA)(DA)(DT)(DA) (DG)(DT)(DT)(DT)(DT)(DT)(DA) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 4.7 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 18000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 4.7 µm / Nominal defocus min: 1.7 µm / Nominal magnification: 18000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-32 / Number real images: 3496 / Average exposure time: 8.0 sec. / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller