[English] 日本語
 Yorodumi
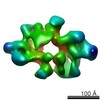
Yorodumi- EMDB-3896: Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3896 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin Complex 2 | |||||||||||||||
 Map data Map data | refined and postprocessed map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationPIP3 activates AKT signaling / CD28 dependent PI3K/Akt signaling / regulation of snRNA pseudouridine synthesis / mitochondria-nucleus signaling pathway / Regulation of TP53 Degradation / positive regulation of Rho guanyl-nucleotide exchange factor activity / TORC2 signaling /  1-phosphatidylinositol 4-kinase / 1-phosphatidylinositol 4-kinase /  1-phosphatidylinositol 4-kinase activity / establishment or maintenance of actin cytoskeleton polarity ...PIP3 activates AKT signaling / CD28 dependent PI3K/Akt signaling / regulation of snRNA pseudouridine synthesis / mitochondria-nucleus signaling pathway / Regulation of TP53 Degradation / positive regulation of Rho guanyl-nucleotide exchange factor activity / TORC2 signaling / 1-phosphatidylinositol 4-kinase activity / establishment or maintenance of actin cytoskeleton polarity ...PIP3 activates AKT signaling / CD28 dependent PI3K/Akt signaling / regulation of snRNA pseudouridine synthesis / mitochondria-nucleus signaling pathway / Regulation of TP53 Degradation / positive regulation of Rho guanyl-nucleotide exchange factor activity / TORC2 signaling /  1-phosphatidylinositol 4-kinase / 1-phosphatidylinositol 4-kinase /  1-phosphatidylinositol 4-kinase activity / establishment or maintenance of actin cytoskeleton polarity / VEGFR2 mediated vascular permeability / HSF1-dependent transactivation / fungal-type cell wall organization / 1-phosphatidylinositol 4-kinase activity / establishment or maintenance of actin cytoskeleton polarity / VEGFR2 mediated vascular permeability / HSF1-dependent transactivation / fungal-type cell wall organization /  TORC2 complex / Amino acids regulate mTORC1 / TORC2 complex / Amino acids regulate mTORC1 /  TORC1 complex / fungal-type vacuole membrane / vacuolar membrane / positive regulation of Rho protein signal transduction / negative regulation of macroautophagy / positive regulation of endocytosis / TORC1 complex / fungal-type vacuole membrane / vacuolar membrane / positive regulation of Rho protein signal transduction / negative regulation of macroautophagy / positive regulation of endocytosis /  TOR signaling / cytoskeleton organization / TOR signaling / cytoskeleton organization /  phosphatidylinositol-4,5-bisphosphate binding / nuclear periphery / negative regulation of autophagy / response to nutrient / phosphatidylinositol-4,5-bisphosphate binding / nuclear periphery / negative regulation of autophagy / response to nutrient /  regulation of cell growth / regulation of actin cytoskeleton organization / regulation of cell growth / regulation of actin cytoskeleton organization /  ribosome biogenesis / endosome membrane / ribosome biogenesis / endosome membrane /  regulation of cell cycle / regulation of cell cycle /  non-specific serine/threonine protein kinase / non-specific serine/threonine protein kinase /  cell cycle / cell cycle /  Golgi membrane / Golgi membrane /  protein phosphorylation / protein serine kinase activity / protein serine/threonine kinase activity / protein-containing complex binding / protein phosphorylation / protein serine kinase activity / protein serine/threonine kinase activity / protein-containing complex binding /  signal transduction / signal transduction /  mitochondrion / mitochondrion /  ATP binding / ATP binding /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||||||||
| Biological species |   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) | |||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 7.9 Å cryo EM / Resolution: 7.9 Å | |||||||||||||||
 Authors Authors | Karuppasamy M / Kusmider B / Oliveira TM / Gaubitz C / Prouteau M / Loewith R / Schaffitzel C | |||||||||||||||
| Funding support | 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2015 Journal: Mol Cell / Year: 2015Title: Molecular Basis of the Rapamycin Insensitivity of Target Of Rapamycin Complex 2. Authors: Christl Gaubitz / Taiana M Oliveira / Manoel Prouteau / Alexander Leitner / Manikandan Karuppasamy / Georgia Konstantinidou / Delphine Rispal / Sandra Eltschinger / Graham C Robinson / ...Authors: Christl Gaubitz / Taiana M Oliveira / Manoel Prouteau / Alexander Leitner / Manikandan Karuppasamy / Georgia Konstantinidou / Delphine Rispal / Sandra Eltschinger / Graham C Robinson / Stéphane Thore / Ruedi Aebersold / Christiane Schaffitzel / Robbie Loewith /    Abstract: Target of Rapamycin (TOR) plays central roles in the regulation of eukaryote growth as the hub of two essential multiprotein complexes: TORC1, which is rapamycin-sensitive, and the lesser ...Target of Rapamycin (TOR) plays central roles in the regulation of eukaryote growth as the hub of two essential multiprotein complexes: TORC1, which is rapamycin-sensitive, and the lesser characterized TORC2, which is not. TORC2 is a key regulator of lipid biosynthesis and Akt-mediated survival signaling. In spite of its importance, its structure and the molecular basis of its rapamycin insensitivity are unknown. Using crosslinking-mass spectrometry and electron microscopy, we determined the architecture of TORC2. TORC2 displays a rhomboid shape with pseudo-2-fold symmetry and a prominent central cavity. Our data indicate that the C-terminal part of Avo3, a subunit unique to TORC2, is close to the FKBP12-rapamycin-binding domain of Tor2. Removal of this sequence generated a FKBP12-rapamycin-sensitive TORC2 variant, which provides a powerful tool for deciphering TORC2 function in vivo. Using this variant, we demonstrate a role for TORC2 in G2/M cell-cycle progression. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3896.map.gz emd_3896.map.gz | 113.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3896-v30.xml emd-3896-v30.xml emd-3896.xml emd-3896.xml | 29.1 KB 29.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3896.png emd_3896.png | 43.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3896 http://ftp.pdbj.org/pub/emdb/structures/EMD-3896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3896 | HTTPS FTP |
-Related structure data
| Related structure data |  6emkMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3896.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3896.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refined and postprocessed map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Target of rapamycin protein complex 2
| Entire | Name: Target of rapamycin protein complex 2 MTOR MTOR |
|---|---|
| Components |
|
-Supramolecule #1: Target of rapamycin protein complex 2
| Supramolecule | Name: Target of rapamycin protein complex 2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Molecular weight | Theoretical: 1.4 MDa |
-Macromolecule #1: Serine/threonine-protein kinase TOR2
| Macromolecule | Name: Serine/threonine-protein kinase TOR2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number:  1-phosphatidylinositol 4-kinase 1-phosphatidylinositol 4-kinase |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) |
| Molecular weight | Theoretical: 281.915438 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Sequence | String: MNKYINKYTT PPNLLSLRQR AEGKHRTRKK LTHKSHSHDD EMSTTSNTDS NHNGPNDSGR VITGSAGHIG KISFVDSELD TTFSTLNLI FDKLKSDVPQ ERASGANELS TTLTSLAREV SAEQFQRFSN SLNNKIFELI HGFTSSEKIG GILAVDTLIS F YLSTEELP ...String: MNKYINKYTT PPNLLSLRQR AEGKHRTRKK LTHKSHSHDD EMSTTSNTDS NHNGPNDSGR VITGSAGHIG KISFVDSELD TTFSTLNLI FDKLKSDVPQ ERASGANELS TTLTSLAREV SAEQFQRFSN SLNNKIFELI HGFTSSEKIG GILAVDTLIS F YLSTEELP NQTSRLANYL RVLIPSSDIE VMRLAANTLG RLTVPGGTLT SDFVEFEVRT CIDWLTLTAD NNSSSSKLEY RR HAALLII KALADNSPYL LYPYVNSILD NIWVPLRDAK LIIRLDAAVA LGKCLTIIQD RDPALGKQWF QRLFQGCTHG LSL NTNDSV HATLLVFREL LSLKAPYLRD KYDDIYKSTM KYKEYKFDVI RREVYAILPL LAAFDPAIFT KKYLDRIMVH YLRY LKNID MNAANNSDKP FILVSIGDIA FEVGSSISPY MTLILDNIRE GLRTKFKVRK QFEKDLFYCI GKLACALGPA FAKHL NKDL LNLMLNCPMS DHMQETLMIL NEKIPSLEST VNSRILNLLS ISLSGEKFIQ SNQYDFNNQF SIEKARKSRN QSFMKK TGE SNDDITDAQI LIQCFKMLQL IHHQYSLTEF VRLITISYIE HEDSSVRKLA ALTSCDLFIK DDICKQTSVH ALHSVSE VL SKLLMIAITD PVAEIRLEIL QHLGSNFDPQ LAQPDNLRLL FMALNDEIFG IQLEAIKIIG RLSSVNPAYV VPSLRKTL L ELLTQLKFSN MPKKKEESAT LLCTLINSSD EVAKPYIDPI LDVILPKCQD ASSAVASTAL KVLGELSVVG GKEMTRYLK ELMPLIINTF QDQSNSFKRD AALTTLGQLA ASSGYVVGPL LDYPELLGIL INILKTENNP HIRRGTVRLI GILGALDPYK HREIEVTSN SKSSVEQNAP SIDIALLMQG VSPSNDEYYP TVVIHNLMKI LNDPSLSIHH TAAIQAIMHI FQNLGLRCVS F LDQIIPGI ILVMRSCPPS QLDFYFQQLG SLISIVKQHI RPHVEKIYGV IREFFPIIKL QITIISVIES ISKALEGEFK RF VPETLTF FLDILENDQS NKRIVPIRIL KSLVTFGPNL EDYSHLIMPI VVRMTEYSAG SLKKISIITL GRLAKNINLS EMS SRIVQA LVRILNNGDR ELTKATMNTL SLLLLQLGTD FVVFVPVINK ALLRNRIQHS VYDQLVNKLL NNECLPTNII FDKE NEVPE RKNYEDEMQV TKLPVNQNIL KNAWYCSQQK TKEDWQEWIR RLSIQLLKES PSACLRSCSS LVSVYYPLAR ELFNA SFSS CWVELQTSYQ EDLIQALCKA LSSSENPPEI YQMLLNLVEF MEHDDKPLPI PIHTLGKYAQ KCHAFAKALH YKEVEF LEE PKNSTIEALI SINNQLHQTD SAIGILKHAQ QHNELQLKET WYEKLQRWED ALAAYNEKEA AGEDSVEVMM GKLRSLY AL GEWEELSKLA SEKWGTAKPE VKKAMAPLAA GAAWGLEQWD EIAQYTSVMK SQSPDKEFYD AILCLHRNNF KKAEVHIF N ARDLLVTELS ALVNESYNRA YNVVVRAQII AELEEIIKYK KLPQNSDKRL TMRETWNTRL LGCQKNIDVW QRILRVRSL VIKPKEDAQV RIKFANLCRK SGRMALAKKV LNTLLEETDD PDHPNTAKAS PPVVYAQLKY LWATGLQDEA LKQLINFTSR MAHDLGLDP NNMIAQSVPQ QSKRVPRHVE DYTKLLARCF LKQGEWRVCL QPKWRLSNPD SILGSYLLAT HFDNTWYKAW H NWALANFE VISMLTSVSK KKQEGSDASS VTDINEFDNG MIGVNTFDAK EVHYSSNLIH RHVIPAIKGF FHSISLSESS SL QDALRLL TLWFTFGGIP EATQAMHEGF NLIQIGTWLE VLPQLISRIH QPNQIVSRSL LSLLSDLGKA HPQALVYPLM VAI KSESLS RQKAALSIIE KMRIHSPVLV DQAELVSHEL IRMAVLWHEQ WYEGLDDASR QFFGEHNTEK MFAALEPLYE MLKR GPETL REISFQNSFG RDLNDAYEWL MNYKKSKDVS NLNQAWDIYY NVFRKIGKQL PQLQTLELQH VSPKLLSAHD LELAV PGTR ASGGKPIVKI SKFEPVFSVI SSKQRPRKFC IKGSDGKDYK YVLKGHEDIR QDSLVMQLFG LVNTLLQNDA ECFRRH LDI QQYPAIPLSP KSGLLGWVPN SDTFHVLIRE HREAKKIPLN IEHWVMLQMA PDYDNLTLLQ KVEVFTYALN NTEGQDL YK VLWLKSRSSE TWLERRTTYT RSLAVMSMTG YILGLGDRHP SNLMLDRITG KVIHIDFGDC FEAAILREKF PEKVPFRL T RMLTYAMEVS GIEGSFRITC ENVMKVLRDN KGSLMAILEA FAFDPLINWG FDLPTKKIEE ETGIQLPVMN ANELLSNGA ITEEEVQRVE NEHKNAIRNA RAMLVLKRIT DKLTGNDIRR FNDLDVPEQV DKLIQQATSV ENLCQHYIGW CPFW |
-Macromolecule #2: Target of rapamycin complex subunit LST8
| Macromolecule | Name: Target of rapamycin complex subunit LST8 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) |
| Molecular weight | Theoretical: 34.077879 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Sequence | String: MSVILVSAGY DHTIRFWEAL TGVCSRTIQH SDSQVNRLEI TNDKKLLATA GHQNVRLYDI RTTNPNPVAS FEGHRGNVTS VSFQQDNRW MVTSSEDGTI KVWDVRSPSI PRNYKHNAPV NEVVIHPNQG ELISCDRDGN IRIWDLGENQ CTHQLTPEDD T SLQSLSMA ...String: MSVILVSAGY DHTIRFWEAL TGVCSRTIQH SDSQVNRLEI TNDKKLLATA GHQNVRLYDI RTTNPNPVAS FEGHRGNVTS VSFQQDNRW MVTSSEDGTI KVWDVRSPSI PRNYKHNAPV NEVVIHPNQG ELISCDRDGN IRIWDLGENQ CTHQLTPEDD T SLQSLSMA SDGSMLAAAN TKGNCYVWEM PNHTDASHLK PVTKFRAHST YITRILLSSD VKHLATCSAD HTARVWSIDD DF KLETTLD GHQRWVWDCA FSADSAYLVT ASSDHYVRLW DLSTREIVRQ YGGHHKGAVC VALNDV |
-Macromolecule #3: Target of rapamycin complex 2 subunit TSC11
| Macromolecule | Name: Target of rapamycin complex 2 subunit TSC11 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) |
| Molecular weight | Theoretical: 25.804662 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
-Macromolecule #4: Target of rapamycin complex 2 subunit AVO2
| Macromolecule | Name: Target of rapamycin complex 2 subunit AVO2 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) |
| Molecular weight | Theoretical: 47.206457 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Sequence | String: MLKEPSVRLR EAIIEGNLLI VKRLLRRNPD LLTNIDSENG WSSLHYASYH GRYLICVYLI QLGHDKHELI KTFKGNTCVH LALMKGHEQ TLHLLLQQFP RFINHRGENG RAPIHIACMN DYYQCLSLLI GVGADLWVMD TNGDTPLHVC LEYGSISCMK M LLNEGEVS ...String: MLKEPSVRLR EAIIEGNLLI VKRLLRRNPD LLTNIDSENG WSSLHYASYH GRYLICVYLI QLGHDKHELI KTFKGNTCVH LALMKGHEQ TLHLLLQQFP RFINHRGENG RAPIHIACMN DYYQCLSLLI GVGADLWVMD TNGDTPLHVC LEYGSISCMK M LLNEGEVS LDDNVRDKGN WKPIDVAQTF EVGNIYSKVL KEVKKKGPPL GAGKKPSSFR TPILNAKATF EDGPSPVLSM NS PYSLYSN NSPLPVLPRR ISTHTTSGNG GNRRSSITNP VFNPRKPTLS TDSFSSSSNS SSRLRVNSIN VKTPVGVSPK KEL VSESVR HSATPTSPHN NIALINRYLL PNKSNDNVRG DSQTATINDD GGGGNGGDAT IGMGLRKDPD DENENKYKIK VNNG EPRRR VSLLNIPISK LRNSNNTRAE D |
-Macromolecule #5: Target of rapamycin complex 2 subunit AVO1
| Macromolecule | Name: Target of rapamycin complex 2 subunit AVO1 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (yeast) |
| Molecular weight | Theoretical: 131.565453 KDa |
| Recombinant expression | Organism:   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast) |
| Sequence | String: MDTVTVLNEL RAQFLRVCPE KDQMKRIIKP YIPVDEFNTE QCLDSSIREL YMNSDGVSLL PELESPPVSK DFMENYASLG KMRIMRENE GQKGKANQNL IGAEKTERDE EETRNLQDKS AKNTLIVEEN GTLRYNPLNS SASNSLLNDD DHTSGKHHKT S SKEDSYLN ...String: MDTVTVLNEL RAQFLRVCPE KDQMKRIIKP YIPVDEFNTE QCLDSSIREL YMNSDGVSLL PELESPPVSK DFMENYASLG KMRIMRENE GQKGKANQNL IGAEKTERDE EETRNLQDKS AKNTLIVEEN GTLRYNPLNS SASNSLLNDD DHTSGKHHKT S SKEDSYLN SSMEMQKKSS KRSSLPFVRI FKSRRDHSNT SGNKNVMNTT NTRAKSSTLH PPGARHNKKG SKFDMNFDFD EN LEEEDDD DDDDEEGDDI HSQFFQLDDD FDAKGSGASP AHKGINGMSN NKNNTYTNNR NSISILDDRE SSNGNIGSAS RLK SHFPTS QKGKIFLTDN KNDGQKSDSL NANKGIHGDG SSASGNGSVS RDGLTETESN NISDMESYIN EKDLDDLNFD TVTS NINKT VSDLGGHEST NDGTAVMNRD SKDSRSNSNE FNAQNRDRIT PGSSYGKSLL GSEYSEERYS NNDSSTMESG EMSLD SDMQ TNTIPSHSIP MSMQKYGIYH GDDDSTLNNV FDKAVLTMNS SRHPKERRDT VISGKEPTSL TSSNRKFSVS SNLTST RSP LLRGHGRTSS TASSEHMKAP KVSDSVLHRA RKSTLTLKQD HSQPSVPSSV HKSSKEGNIL IEKTTDYLVS KPKASQL SN MFNKKKKRTN TNSVDVLEYF SFVCGDKVPN YESMGLEIYI QASKKYKRNS FTTKVRKSST IFEVIGFALF LYSTEKKP D NFEEDGLTVE DISNPNNFSL KIVDEDGEPF EDNFGKLDRK STIQSISDSE VVLCKVDDAE KSQNEIETPL PFETGGGLM DASTLDANSS HDTTDGTINQ LSFYKPIIGN EDDIDKTNGS KIIDVTVYLY PNVNPKFNYT TISVLVTSHI NDILVKYCKM KNMDPNEYA LKVLGKNYIL DLNDTVLRLD GINKVELISK KDARELHLEK MKPDLKKPVL PTIQSNDLTP LTLEPLNSYL K ADAGGAVA AIPENTKVTS KAKKISTKYK LGLAKQHSSS SVASGSVSTA GGLANGNGFF KNKNSSKSSL HGTLQFHNIN RS QSTMEHT PDTPNGVGDN FQDLFTGAYH KYKVWRRQQM SFINKHERTL AIDGDYIYIV PPEGRIHWHD NVKTKSLHIS QVV LVKKSK RVPEHFKIFV RREGQDDIKR YYFEAVSGQE CTEIVTRLQN LLSAYRMNHK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.01 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV / Details: 2 - 3 sec blotting. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: FEI FALCON II (4k x 4k) / #0 - Detector mode: INTEGRATING / #0 - Number grids imaged: 4 / #0 - Number real images: 4189 / #0 - Average exposure time: 2.3 sec. / #0 - Average electron dose: 50.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 QUANTUM (4k x 4k) / #1 - Detector mode: SUPER-RESOLUTION / #1 - Digitization - Dimensions - Width: 3710 pixel / #1 - Digitization - Dimensions - Height: 3838 pixel / #1 - Digitization - Frames/image: 1-40 / #1 - Number grids imaged: 1 / #1 - Number real images: 2847 / #1 - Average exposure time: 20.0 sec. / #1 - Average electron dose: 47.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing #1
Image processing #1
| Particle selection | Number selected: 350000 |
|---|---|
| CTF correction | Software - Name: CTFFIND (ver. 4) |
| Startup model | Type of model: EMDB MAP EMDB ID: |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 1.4) |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 1.4) |
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 7.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 16190 ) / Resolution.type: BY AUTHOR / Resolution: 7.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 16190 |
| Image processing ID | 1 |
| Image recording ID | 1 |
- Image processing #2
Image processing #2
| Particle selection | Number selected: 200000 |
|---|---|
| CTF correction | Software - Name: CTFFIND (ver. 4) |
| Startup model | Type of model: EMDB MAP EMDB ID: |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 1.4) |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 1.4) |
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 8.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 10663 ) / Resolution.type: BY AUTHOR / Resolution: 8.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 10663 |
| Image processing ID | 2 |
| Image recording ID | 2 |
 Movie
Movie Controller
Controller