[English] 日本語
 Yorodumi
Yorodumi- EMDB-3818: Structure of an RNA polymerase II-DSIF transcription elongation c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3818 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of an RNA polymerase II-DSIF transcription elongation complex, DSIF-EC4 | ||||||||||||
 Map data Map data | Locally filtered map, B-factor 0 | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |   Bos taurus (cattle) / Bos taurus (cattle) /   Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Bernecky C / Plitzko JM / Cramer P | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2017 Journal: Nat Struct Mol Biol / Year: 2017Title: Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Authors: Carrie Bernecky / Jürgen M Plitzko / Patrick Cramer /  Abstract: During transcription, RNA polymerase II (Pol II) associates with the conserved elongation factor DSIF. DSIF renders the elongation complex stable and functions during Pol II pausing and RNA ...During transcription, RNA polymerase II (Pol II) associates with the conserved elongation factor DSIF. DSIF renders the elongation complex stable and functions during Pol II pausing and RNA processing. We combined cryo-EM and X-ray crystallography to determine the structure of the mammalian Pol II-DSIF elongation complex at a nominal resolution of 3.4 Å. Human DSIF has a modular structure with two domains forming a DNA clamp, two domains forming an RNA clamp, and one domain buttressing the RNA clamp. The clamps maintain the transcription bubble, position upstream DNA, and retain the RNA transcript in the exit tunnel. The mobile C-terminal region of DSIF is located near exiting RNA, where it can recruit factors for RNA processing. The structure provides insight into the roles of DSIF during mRNA synthesis. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3818.map.gz emd_3818.map.gz | 58.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3818-v30.xml emd-3818-v30.xml emd-3818.xml emd-3818.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3818.png emd_3818.png | 102.5 KB | ||
| Masks |  emd_3818_msk_1.map emd_3818_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_3818_additional_1.map.gz emd_3818_additional_1.map.gz emd_3818_additional_2.map.gz emd_3818_additional_2.map.gz emd_3818_half_map_1.map.gz emd_3818_half_map_1.map.gz emd_3818_half_map_2.map.gz emd_3818_half_map_2.map.gz | 59.9 MB 59.7 MB 49.7 MB 49.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3818 http://ftp.pdbj.org/pub/emdb/structures/EMD-3818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3818 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3818 | HTTPS FTP |
-Related structure data
| Related structure data |  3815C  3816C  3817C  3819C  5ohoC  5ohqC  5oikC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3818.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3818.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally filtered map, B-factor 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_3818_msk_1.map emd_3818_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
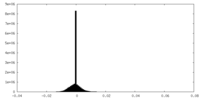
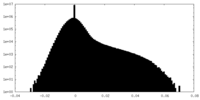
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: None
| File | emd_3818_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: None
| File | emd_3818_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap 1
| File | emd_3818_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Halfmap 2
| File | emd_3818_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RNA polymerase II-DSIF elongation complex
| Entire | Name: RNA polymerase II-DSIF elongation complex |
|---|---|
| Components |
|
-Supramolecule #1: RNA polymerase II-DSIF elongation complex
| Supramolecule | Name: RNA polymerase II-DSIF elongation complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 690 KDa |
-Supramolecule #2: RNA polymerase II
| Supramolecule | Name: RNA polymerase II / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Bos taurus (cattle) Bos taurus (cattle) |
| Molecular weight | Theoretical: 520 KDa |
-Supramolecule #3: DSIF
| Supramolecule | Name: DSIF / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Molecular weight | Theoretical: 130 KDa |
-Supramolecule #4: DNA-RNA synthetic construct
| Supramolecule | Name: DNA-RNA synthetic construct / type: complex / ID: 4 / Parent: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 40 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.25 Component:
Details: pH was adjusted at 25 degrees Celsius | |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Sample (four microliters) was applied to glow-discharged Quantifoil R 2/1 holey carbon grids, which were then blotted for 8.5s and plunge-frozen in liquid ethane.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.6 µm / Calibrated defocus min: 0.6 µm / Calibrated magnification: 37037 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 37000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.6 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 37000 |
| Specialist optics | Energy filter - Name: GIF Quantum / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-33 / Number grids imaged: 1 / Number real images: 2549 / Average exposure time: 9.9 sec. / Average electron dose: 33.0 e/Å2 Details: Movies were aligned and binned to the physical pixel size of 1.35 angstroms. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 687928 |
|---|---|
| CTF correction | Software - Name: CTFFIND (ver. 4.0.16) |
| Startup model | Type of model: EMDB MAP EMDB ID: |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 1.4) |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 1.4) |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 76394 |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X