+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3785 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Watermelon mosaic virus potyvirus. | |||||||||
 Map data Map data | Cryoelectron microscopy map of Watermelon mosaic virus filament. Resulting map from Relion postprocessing. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Potyvirus coat protein / Potyvirus coat protein /  viral capsid / Coat protein viral capsid / Coat protein Function and homology information Function and homology information | |||||||||
| Biological species |   Watermelon mosaic virus Watermelon mosaic virus | |||||||||
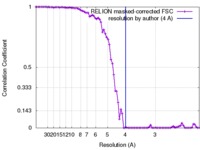
| Method | helical reconstruction /  cryo EM / Resolution: 4.0 Å cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Zamora M / Mendez-Lopez E / Agirrezabala X / Cuesta R / Lavin JL / Sanchez-Pina MA / Aranda M / Valle M | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2017 Journal: Sci Adv / Year: 2017Title: Potyvirus virion structure shows conserved protein fold and RNA binding site in ssRNA viruses. Authors: Miguel Zamora / Eduardo Méndez-López / Xabier Agirrezabala / Rebeca Cuesta / José L Lavín / M Amelia Sánchez-Pina / Miguel A Aranda / Mikel Valle /  Abstract: Potyviruses constitute the second largest genus of plant viruses and cause important economic losses in a large variety of crops; however, the atomic structure of their particles remains unknown. ...Potyviruses constitute the second largest genus of plant viruses and cause important economic losses in a large variety of crops; however, the atomic structure of their particles remains unknown. Infective potyvirus virions are long flexuous filaments where coat protein (CP) subunits assemble in helical mode bound to a monopartite positive-sense single-stranded RNA [(+)ssRNA] genome. We present the cryo-electron microscopy (cryoEM) structure of the potyvirus watermelon mosaic virus at a resolution of 4.0 Å. The atomic model shows a conserved fold for the CPs of flexible filamentous plant viruses, including a universally conserved RNA binding pocket, which is a potential target for antiviral compounds. This conserved fold of the CP is widely distributed in eukaryotic viruses and is also shared by nucleoproteins of enveloped viruses with segmented (-)ssRNA (negative-sense ssRNA) genomes, including influenza viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3785.map.gz emd_3785.map.gz | 42.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3785-v30.xml emd-3785-v30.xml emd-3785.xml emd-3785.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3785_fsc.xml emd_3785_fsc.xml | 7.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_3785.png emd_3785.png | 245.2 KB | ||
| Others |  emd_3785_additional.map.gz emd_3785_additional.map.gz emd_3785_half_map_1.map.gz emd_3785_half_map_1.map.gz emd_3785_half_map_2.map.gz emd_3785_half_map_2.map.gz | 17.4 MB 35.5 MB 35.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3785 http://ftp.pdbj.org/pub/emdb/structures/EMD-3785 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3785 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3785 | HTTPS FTP |
-Related structure data
| Related structure data |  5odvMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3785.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3785.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryoelectron microscopy map of Watermelon mosaic virus filament. Resulting map from Relion postprocessing. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Cryoelectron microscopy map of Watermelon mosaic virus filament....
| File | emd_3785_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryoelectron microscopy map of Watermelon mosaic virus filament. Map generated by applying helical symmetry parameters to Relion postprocessing resulting map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half 1 map of Watermelon mosaic virus...
| File | emd_3785_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half 1 map of Watermelon mosaic virus filament resulting from Relion 3D Refine. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half 2 map of Watermelon mosaic virus...
| File | emd_3785_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half 2 map of Watermelon mosaic virus filament resulting from Relion 3D Refine. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Watermelon mosaic virus
| Entire | Name:   Watermelon mosaic virus Watermelon mosaic virus |
|---|---|
| Components |
|
-Supramolecule #1: Watermelon mosaic virus
| Supramolecule | Name: Watermelon mosaic virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 146500 / Sci species name: Watermelon mosaic virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: coat protein
| Macromolecule | Name: coat protein / type: protein_or_peptide / ID: 1 Details: 57 residues from N-terminus and 17 residues from C-terminus are not present in our pdb model due to the absence of densities for them in the cryo-electron microscopy map as a consequence of ...Details: 57 residues from N-terminus and 17 residues from C-terminus are not present in our pdb model due to the absence of densities for them in the cryo-electron microscopy map as a consequence of being high flexible regions in the protein. Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Watermelon mosaic virus Watermelon mosaic virus |
| Molecular weight | Theoretical: 31.463412 KDa |
| Sequence | String: SGKEAVENLD AGKESKKDTS GKGDKPQNSQ TGQGSKEQTK TGTVSKDVNV GSKGKEVPRL QKITKKMNLP TVGGKIILSL DHLLEYKPN QVDLFNTRAT KTQFESWYSA VKVEYDLNDE QMGVIMNGFM VWCIDNGTSP DVNGVWVMMD GEEQVEYPLK P IVENAKPT ...String: SGKEAVENLD AGKESKKDTS GKGDKPQNSQ TGQGSKEQTK TGTVSKDVNV GSKGKEVPRL QKITKKMNLP TVGGKIILSL DHLLEYKPN QVDLFNTRAT KTQFESWYSA VKVEYDLNDE QMGVIMNGFM VWCIDNGTSP DVNGVWVMMD GEEQVEYPLK P IVENAKPT LRQIMHHFSD AAEAYIEMRN SESPYMPRYG LLRNLRDREL ARYAFDFYEV TSKTPNRARE AIAQMKAAAL AG INSRLFG LDGNISTNSE NTERHTARDV NQNMHTLLGM GPPQ |
-Macromolecule #2: RNA (5'-R(P*UP*UP*UP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*UP*UP*UP*U)-3') / type: rna / ID: 2 / Number of copies: 24 |
|---|---|
| Source (natural) | Organism:   Watermelon mosaic virus Watermelon mosaic virus |
| Molecular weight | Theoretical: 1.485872 KDa |
| Sequence | String: UUUUU |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Frames/image: 2-27 / Average electron dose: 1.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X