[English] 日本語
 Yorodumi
Yorodumi- EMDB-7089: HIV-1 Envelope SOSIP trimer clone PC64M4c054 in complex with auto... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7089 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 Envelope SOSIP trimer clone PC64M4c054 in complex with autologous PCT64-13C Fab at 13.2 A resolution | |||||||||
 Map data Map data | Unsharpened map from Relion 3D auto-refinement filtered at FSC=0.143 resolution | |||||||||
 Sample Sample |
| |||||||||
| Biological species |    Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 13.2 Å cryo EM / Resolution: 13.2 Å | |||||||||
 Authors Authors | Rantalainen K / Berndsen ZT / Ward AB | |||||||||
 Citation Citation |  Journal: Immunity / Year: 2017 Journal: Immunity / Year: 2017Title: HIV Envelope Glycoform Heterogeneity and Localized Diversity Govern the Initiation and Maturation of a V2 Apex Broadly Neutralizing Antibody Lineage. Authors: Elise Landais / Ben Murrell / Bryan Briney / Sasha Murrell / Kimmo Rantalainen / Zachary T Berndsen / Alejandra Ramos / Lalinda Wickramasinghe / Melissa Laird Smith / Kemal Eren / Natalia de ...Authors: Elise Landais / Ben Murrell / Bryan Briney / Sasha Murrell / Kimmo Rantalainen / Zachary T Berndsen / Alejandra Ramos / Lalinda Wickramasinghe / Melissa Laird Smith / Kemal Eren / Natalia de Val / Mengyu Wu / Audrey Cappelletti / Jeffrey Umotoy / Yolanda Lie / Terri Wrin / Paul Algate / Po-Ying Chan-Hui / Etienne Karita / / / Andrew B Ward / Ian A Wilson / Dennis R Burton / Davey Smith / Sergei L Kosakovsky Pond / Pascal Poignard /    Abstract: Understanding how broadly neutralizing antibodies (bnAbs) to HIV envelope (Env) develop during natural infection can help guide the rational design of an HIV vaccine. Here, we described a bnAb ...Understanding how broadly neutralizing antibodies (bnAbs) to HIV envelope (Env) develop during natural infection can help guide the rational design of an HIV vaccine. Here, we described a bnAb lineage targeting the Env V2 apex and the Ab-Env co-evolution that led to development of neutralization breadth. The lineage Abs bore an anionic heavy chain complementarity-determining region 3 (CDRH3) of 25 amino acids, among the shortest known for this class of Abs, and achieved breadth with only 10% nucleotide somatic hypermutation and no insertions or deletions. The data suggested a role for Env glycoform heterogeneity in the activation of the lineage germline B cell. Finally, we showed that localized diversity at key V2 epitope residues drove bnAb maturation toward breadth, mirroring the Env evolution pattern described for another donor who developed V2-apex targeting bnAbs. Overall, these findings suggest potential strategies for vaccine approaches based on germline-targeting and serial immunogen design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7089.map.gz emd_7089.map.gz | 97.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7089-v30.xml emd-7089-v30.xml emd-7089.xml emd-7089.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7089.png emd_7089.png | 64.2 KB | ||
| Masks |  emd_7089_msk_1.map emd_7089_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_7089_half_map_1.map.gz emd_7089_half_map_1.map.gz emd_7089_half_map_2.map.gz emd_7089_half_map_2.map.gz | 98.3 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7089 http://ftp.pdbj.org/pub/emdb/structures/EMD-7089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7089 | HTTPS FTP |
-Related structure data
| Related structure data |  7104C  7105C  7106C  7107C  7108C  5fehC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7089.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7089.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map from Relion 3D auto-refinement filtered at FSC=0.143 resolution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_7089_msk_1.map emd_7089_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: unfiltered half map from Relion 3D auto-refinement
| File | emd_7089_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half map from Relion 3D auto-refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: unfiltered half map from Relion 3D auto-refinement
| File | emd_7089_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half map from Relion 3D auto-refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 Envelope Glycoprotein SOSIP.664 clone PC64M4c054 in complex...
| Entire | Name: HIV-1 Envelope Glycoprotein SOSIP.664 clone PC64M4c054 in complex with autologous Fab PCT64-13C |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 Envelope Glycoprotein SOSIP.664 clone PC64M4c054 in complex...
| Supramolecule | Name: HIV-1 Envelope Glycoprotein SOSIP.664 clone PC64M4c054 in complex with autologous Fab PCT64-13C type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Fab generated by proteolytic cleavage of IgG antibody |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 275 KDa |
-Macromolecule #1: HIV-1 Envelope Glycoprotein SOSIP.664 clone PC64M4c054
| Macromolecule | Name: HIV-1 Envelope Glycoprotein SOSIP.664 clone PC64M4c054 type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:    Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARANNLW VTVYYGVPVW RDAETTLFCA SDAKAYDTEV HNVWATHACV PTDPSPQEIH LANVTEKFNM WKNSMVEQMH TDIISLWDES LKPCVKLTPL CITLNCTNIT RKNVTGGNLT EDGKEELKNC SFNATTELRN ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARANNLW VTVYYGVPVW RDAETTLFCA SDAKAYDTEV HNVWATHACV PTDPSPQEIH LANVTEKFNM WKNSMVEQMH TDIISLWDES LKPCVKLTPL CITLNCTNIT RKNVTGGNLT EDGKEELKNC SFNATTELRN KRQKVHSLFY RLDLVELNEG NSSNSNTSMY RLINCNTSAI TQACPKVSFE PIPIHYCAPA GFAILKCREE EFNGTGPCKN VSTVQCTHGI KPVVSTQLLL NGSLAEGTVK IRCENISNNA KTILVQLTTP VRINCTRPNN NTRTSIRIGP GQSFYATGDI IGDIRKAYCN VSGSEWKEAL GKVVVQLRSH FNKTITFASS SGGDLEITTH SFNCGGEFFY CNTSSLFNST WDGNSTTNST QEPNGTITLP CRIKQIINMW QRTGQAMYAP PIPGKIRCDS NITGLILTRD GENNNTESET FRPEGGDMRN NWRSELYKYK VVKIDPLGVA PTGCKRRVVE RRRRRRAVGI GAVLFGFLGA AGSTMGAASL TLTVQARQLL SGIVQQQSNL LRAPEAQQHL LRLTVWGIKQ LQARVLAVER YLSDQQLLGI WGCSGKLICC TTVPWNSSWS NKSQDEIWNN MTWLQWDKEI SNYTDTIYYL IEKSQNQQEV NEKDLLALD |
-Macromolecule #2: Immunoglobulin G PCT64-13C Heavy Chain
| Macromolecule | Name: Immunoglobulin G PCT64-13C Heavy Chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVKPGGSLRL SCAASGFTFT NAWLDWVRQA PGKGLEWVGR IKSKTDGGTT DHAAPVKGRF TISRDDSKNT VYLQMNSLKI EDTAVYYCTT GVETYDFWSG YDDHYYDYYF KDVWGKGTTV TVSS |
-Macromolecule #3: Immunoglobulin G PCT64-13C Light Chain
| Macromolecule | Name: Immunoglobulin G PCT64-13C Light Chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQSVS NNYLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSG TDFTLTISRL EPEDFAVYYC QQSARSFTFG PGTKVDIK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: TBS + DDM (added to a final concentration of 0.06 mM prior to vitrification) |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.7794 µm / Nominal defocus min: 0.8687 µm / Nominal magnification: 29000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.7794 µm / Nominal defocus min: 0.8687 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-43 / Number grids imaged: 1 / Number real images: 100 / Average exposure time: 0.2 sec. / Average electron dose: 2.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 12343 |
|---|---|
| CTF correction | Software - Name: Gctf (ver. 1.06) |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 2.0) |
| Final 3D classification | Software - Name: RELION (ver. 2.0) |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 2.0) |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 13.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 2.0) / Number images used: 2651 |
 Movie
Movie Controller
Controller



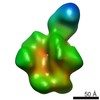







 Z
Z Y
Y X
X