[English] 日本語
 Yorodumi
Yorodumi- EMDB-1596: HIV-1 Env spike maps using various alignment and classfication co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1596 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 Env spike maps using various alignment and classfication combinations | |||||||||
 Map data Map data | Classified HIV-1 Env spikes using different alignment and classification strategies. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  HIV-1 / envelope spike HIV-1 / envelope spike | |||||||||
| Biological species |    Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
| Method | subtomogram averaging /  cryo EM cryo EM | |||||||||
 Authors Authors | Zhu P / Winkler H / Chertova E / Taylor KA / Roux KH | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2008 Journal: PLoS Pathog / Year: 2008Title: Cryoelectron tomography of HIV-1 envelope spikes: further evidence for tripod-like legs. Authors: Ping Zhu / Hanspeter Winkler / Elena Chertova / Kenneth A Taylor / Kenneth H Roux /  Abstract: A detailed understanding of the morphology of the HIV-1 envelope (Env) spike is key to understanding viral pathogenesis and for informed vaccine design. We have previously presented a cryoelectron ...A detailed understanding of the morphology of the HIV-1 envelope (Env) spike is key to understanding viral pathogenesis and for informed vaccine design. We have previously presented a cryoelectron microscopic tomogram (cryoET) of the Env spikes on SIV virions. Several structural features were noted in the gp120 head and gp41 stalk regions. Perhaps most notable was the presence of three splayed legs projecting obliquely from the base of the spike head toward the viral membrane. Subsequently, a second 3D image of SIV spikes, also obtained by cryoET, was published by another group which featured a compact vertical stalk. We now report the cryoET analysis of HIV-1 virion-associated Env spikes using enhanced analytical cryoET procedures. More than 2,000 Env spike volumes were initially selected, aligned, and sorted into structural classes using algorithms that compensate for the "missing wedge" and do not impose any symmetry. The results show varying morphologies between structural classes: some classes showed trimers in the head domains; nearly all showed two or three legs, though unambiguous three-fold symmetry was not observed either in the heads or the legs. Subsequently, clearer evidence of trimeric head domains and three splayed legs emerged when head and leg volumes were independently aligned and classified. These data show that HIV-1, like SIV, also displays the tripod-like leg configuration, and, unexpectedly, shows considerable gp41 leg flexibility/heteromorphology. The tripod-like model for gp41 is consistent with, and helps explain, many of the unique biophysical and immunological features of this region. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1596.map.gz emd_1596.map.gz | 931.5 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1596-v30.xml emd-1596-v30.xml emd-1596.xml emd-1596.xml | 8.2 KB 8.2 KB | Display Display |  EMDB header EMDB header |
| Images |  1596.gif 1596.gif | 49.2 KB | ||
| Others |  1596_class_maps.zip 1596_class_maps.zip Schematic.jpg Schematic.jpg readme.txt readme.txt | 36.4 MB 17.8 KB 779 B | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1596 http://ftp.pdbj.org/pub/emdb/structures/EMD-1596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1596 | HTTPS FTP |
-Related structure data
| Related structure data |  1513C  1514C  1515C  1516C  1517C  1518C  1519C  1520C  1521C  1522C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1596.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1596.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Classified HIV-1 Env spikes using different alignment and classification strategies. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.56 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
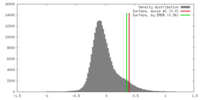
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Others
| Details | [readme.txt] Reference: Zhu P, Winkler H, Chertova E, Taylor KA, Roux KH (2008) Cryoelectron Tomography of HIV-1 Envelope Spikes: Further Evidence for Tripod-Like Legs. PLoS Pathog 4(11): e1000203. The folders in 1596_class_maps.zip consist of aligned and classified HIV-1 Env spikes maps using various alignment and classification combinations as shown in figure 3 of the reference. Please refer back to the reference for details. A_WW: Panel A Aligned: whole, Classified: whole B_WH: Panel B Aligned: whole, Classified: head C_WL: Panel C Aligned: whole, Classified: legs D_HH: Panel D Aligned: head, Classified: head E_LL: Panel E Aligned: legs, Classified: legs In each subfolder, *-cls-[1-8].map: eight classes produced for each combination |
|---|---|
| Image | |
| File | 1596_class_maps.zip (36.4 MB) |
- Sample components
Sample components
-Entire : HIV-1 BaL / SUPT1-CCR5 CL.30, provided by the AIDS Vaccine Progra...
| Entire | Name: HIV-1 BaL / SUPT1-CCR5 CL.30, provided by the AIDS Vaccine Program, SAIC Frederick, Inc., NCI, Frederick, MD. Lot p3955. |
|---|---|
| Components |
|
-Supramolecule #1000: HIV-1 BaL / SUPT1-CCR5 CL.30, provided by the AIDS Vaccine Progra...
| Supramolecule | Name: HIV-1 BaL / SUPT1-CCR5 CL.30, provided by the AIDS Vaccine Program, SAIC Frederick, Inc., NCI, Frederick, MD. Lot p3955. type: sample / ID: 1000 Details: The envelope spikes are distributed on the surface of HIV-1 virions. Oligomeric state: Highly purified virus. The envelope spikes on the virion surface are trimer. Number unique components: 1 |
|---|
-Supramolecule #1: Human immunodeficiency virus 1
| Supramolecule | Name: Human immunodeficiency virus 1 / type: virus / ID: 1 / Name.synonym: HIV-1 BaL SUPT1-CCR5 CL.30 Details: The envelope spikes are distributed on the surface of HIV-1 virions. NCBI-ID: 11676 / Sci species name: Human immunodeficiency virus 1 / Database: NCBI / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No / Syn species name: HIV-1 BaL SUPT1-CCR5 CL.30 |
|---|---|
| Host (natural) | Organism:   Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
- Sample preparation
Sample preparation
| Concentration | 2.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: Originally in TNE, resuspend in PBS |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: Home made. Vitrification carried out in cold room Method: Blot for 5-6 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal magnification: 43200 Bright-field microscopy / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal magnification: 43200 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Image recording | Average electron dose: 1.5 e/Å2 |
- Image processing
Image processing
| Final reconstruction | Software - Name: Protomo |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)