[English] 日本語
 Yorodumi
Yorodumi- EMDB-9277: Single particle cryoEM structure of a DARPin-aldolase platform in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9277 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle cryoEM structure of a DARPin-aldolase platform in complex with GFP | |||||||||
 Map data Map data | DARPin-aldolase platform in complex with GFP, Relion Post processed map with B factor -25 angstrom squared | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | synthetic construct / platform / single particle cryoEM / small protein display / LYASE-FLUORESCENT PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of Arp2/3 complex-mediated actin nucleation /  fructose-bisphosphate aldolase / fructose-bisphosphate aldolase /  fructose-bisphosphate aldolase activity / fructose-bisphosphate aldolase activity /  M band / I band / M band / I band /  bioluminescence / generation of precursor metabolites and energy / glycolytic process / protein homotetramerization / positive regulation of cell migration bioluminescence / generation of precursor metabolites and energy / glycolytic process / protein homotetramerization / positive regulation of cell migrationSimilarity search - Function | |||||||||
| Biological species | synthetic construct (others) /   Aequorea victoria (jellyfish) / Aequorea victoria (jellyfish) /   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Weaver SJ / Yao Q | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2019 Journal: Structure / Year: 2019Title: Fusion of DARPin to Aldolase Enables Visualization of Small Protein by Cryo-EM. Authors: Qing Yao / Sara J Weaver / Jee-Young Mock / Grant J Jensen /  Abstract: Solving protein structures by single-particle cryoelectron microscopy (cryo-EM) has become a crucial tool in structural biology. While exciting progress is being made toward the visualization of ...Solving protein structures by single-particle cryoelectron microscopy (cryo-EM) has become a crucial tool in structural biology. While exciting progress is being made toward the visualization of small macromolecules, the median protein size in both eukaryotes and bacteria is still beyond the reach of cryo-EM. To overcome this problem, we implemented a platform strategy in which a small protein target was rigidly attached to a large, symmetric base via a selectable adapter. Of our seven designs, the best construct used a designed ankyrin repeat protein (DARPin) rigidly fused to tetrameric rabbit muscle aldolase through a helical linker. The DARPin retained its ability to bind its target: GFP. We solved the structure of this complex to 3.0 Å resolution overall, with 5-8 Å resolution in the GFP region. As flexibility in the DARPin position limited the overall resolution of the target, we describe strategies to rigidify this element. #1:  Journal: Biorxiv / Year: 2018 Journal: Biorxiv / Year: 2018Title: Fusion of DARPin to aldolase enables visualization of small protein by cryoEM Authors: Yao Q / Weaver SJ / Mock JY / Jensen GJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9277.map.gz emd_9277.map.gz | 14.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9277-v30.xml emd-9277-v30.xml emd-9277.xml emd-9277.xml | 24.2 KB 24.2 KB | Display Display |  EMDB header EMDB header |
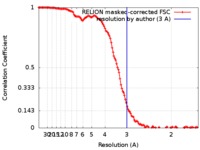
| FSC (resolution estimation) |  emd_9277_fsc.xml emd_9277_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_9277.png emd_9277.png | 67.9 KB | ||
| Filedesc metadata |  emd-9277.cif.gz emd-9277.cif.gz | 7.5 KB | ||
| Others |  emd_9277_half_map_1.map.gz emd_9277_half_map_1.map.gz emd_9277_half_map_2.map.gz emd_9277_half_map_2.map.gz | 140.8 MB 140.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9277 http://ftp.pdbj.org/pub/emdb/structures/EMD-9277 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9277 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9277 | HTTPS FTP |
-Related structure data
| Related structure data |  6mwqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9277.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9277.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DARPin-aldolase platform in complex with GFP, Relion Post processed map with B factor -25 angstrom squared | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8315 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 1 of a DARPin-aldolase platform in complex with GFP
| File | emd_9277_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of a DARPin-aldolase platform in complex with GFP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 of a DARPin-aldolase platform in complex with GFP
| File | emd_9277_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of a DARPin-aldolase platform in complex with GFP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DARPin-aldolase platform in complex with GFP
| Entire | Name: DARPin-aldolase platform in complex with GFP |
|---|---|
| Components |
|
-Supramolecule #1: DARPin-aldolase platform in complex with GFP
| Supramolecule | Name: DARPin-aldolase platform in complex with GFP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 314 KDa |
-Supramolecule #2: DARPin, Muscle-type aldolase chimeric fusion
| Supramolecule | Name: DARPin, Muscle-type aldolase chimeric fusion / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Aequorea victoria (jellyfish) Aequorea victoria (jellyfish) |
-Supramolecule #3: Green fluorescent protein
| Supramolecule | Name: Green fluorescent protein / type: complex / ID: 3 / Parent: 1 |
|---|
-Macromolecule #1: DARPin, Muscle-type aldolase chimeric fusion
| Macromolecule | Name: DARPin, Muscle-type aldolase chimeric fusion / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number:  fructose-bisphosphate aldolase fructose-bisphosphate aldolase |
|---|---|
| Source (natural) | Organism:   Oryctolagus cuniculus (rabbit) Oryctolagus cuniculus (rabbit) |
| Molecular weight | Theoretical: 53.113434 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: SGSDLGKKLL EAARAGQDDE VRILMANGAD VNALDRFGLT PLHLAAQRGH LEIVEVLLKC GADVNAADLW GQTPLHLAAT AGHLEIVEV LLKYGADVNA LDLIGKTPLH LTAIDGHLEI VEVLLKHGAD VNAQDKFGKT AFDISIDNGN EDLAEILQKL N LSDIAHRI ...String: SGSDLGKKLL EAARAGQDDE VRILMANGAD VNALDRFGLT PLHLAAQRGH LEIVEVLLKC GADVNAADLW GQTPLHLAAT AGHLEIVEV LLKYGADVNA LDLIGKTPLH LTAIDGHLEI VEVLLKHGAD VNAQDKFGKT AFDISIDNGN EDLAEILQKL N LSDIAHRI VAPGKGILAA DESTGSIAKR LQSIGTENTE ENRRFYRQLL LTADDRVNPC IGGVILFHET LYQKADDGRP FP QVIKSKG GVVGIKVDKG VVPLAGTNGE TTTQGLDGLS ERCAQYKKDG ADFAKWRCVL KIGEHTPSAL AIMENANVLA RYA SICQQN GIVPIVEPEI LPDGDHDLKR CQYVTEKVLA AVYKALSDHH IYLEGTLLKP NMVTPGHACT QKYSHEEIAM ATVT ALRRT VPPAVTGVTF LSGGQSEEEA SINLNAINKC PLLKPWALTF SYGRALQASA LKAWGGKKEN LKAAQEEYVK RALAN SLAC QGKYTPSGQ UniProtKB: Fructose-bisphosphate aldolase A |
-Macromolecule #2: Green fluorescent protein
| Macromolecule | Name: Green fluorescent protein / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aequorea victoria (jellyfish) Aequorea victoria (jellyfish) |
| Molecular weight | Theoretical: 25.473697 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: SKGEELFTGV VPILVELDGD VNGHKFSVSG EGEGDATYGK LTLKFICTTG KLPVPWPTLV TTLVQCFSRY PDHMKQHDFF KSAMPEGYV QERTIFFKDD GNYKTRAEVK FEGDTLVNRI ELKGIDFKED GNILGHKLEY NYNSHNVYIM ADKQKNGIKV N FKIRHNIE ...String: SKGEELFTGV VPILVELDGD VNGHKFSVSG EGEGDATYGK LTLKFICTTG KLPVPWPTLV TTLVQCFSRY PDHMKQHDFF KSAMPEGYV QERTIFFKDD GNYKTRAEVK FEGDTLVNRI ELKGIDFKED GNILGHKLEY NYNSHNVYIM ADKQKNGIKV N FKIRHNIE DGSVQLADHY QQNTPIGDGP VLLPDNHYLS TQSALSKDPN EKRDHMVLLE FVTAAGIT UniProtKB:  Green fluorescent protein Green fluorescent protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER Details: Grids were frozen on a manual plunger at the Scripps Research Institute Core Microscopy Facility in a 4 degrees C cold room humidified to >95%.. | |||||||||
| Details | The protein complex was then purified with Ni-NTA affinity chromatography (Qiagen), and Superdex 200 chromatography (GE healthcare). The purified GFP-DARPin-aldolase complex was concentrated to 2.5mg/ml in a buffer containing 25 mM Tris-HCl pH 8.0 and 150 mM NaCl. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 30.0 µm / Nominal defocus min: 10.0 µm / Nominal magnification: 165000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 30.0 µm / Nominal defocus min: 10.0 µm / Nominal magnification: 165000 |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #0 - Detector mode: SUPER-RESOLUTION / #0 - Number grids imaged: 1 / #0 - Number real images: 1133 / #0 - Average exposure time: 4.0 sec. / #0 - Average electron dose: 2.3 e/Å2 #0 - Details: All three microscope sessions used grids frozen in the same session. In the first microscopy session the stage was untilted (0 degrees). #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #1 - Detector mode: SUPER-RESOLUTION / #1 - Number grids imaged: 1 / #1 - Number real images: 548 / #1 - Average exposure time: 4.0 sec. / #1 - Average electron dose: 2.3 e/Å2 #1 - Details: All three microscope sessions used grids frozen in the same session. In the second microscopy session the stage was untilted (0 degrees). #2 - Image recording ID: 3 / #2 - Film or detector model: GATAN K2 SUMMIT (4k x 4k) / #2 - Detector mode: SUPER-RESOLUTION / #2 - Digitization - Frames/image: 1-40 / #2 - Number grids imaged: 1 / #2 - Number real images: 1180 / #2 - Average exposure time: 4.0 sec. / #2 - Average electron dose: 1.1 e/Å2 #2 - Details: All three microscope sessions used grids frozen in the same session. In the third microscopy session the stage was tilted to 26 degrees. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | PDB models 5vy5 and 5ma6 were used as starting points. These models were mutated to match the sequence of the DARPin-aldolase platform in complex with GFP that were used. The PDB models were docked into the cryoEM density using Chimera fit into map function. No model refinement was used. | ||||||||
| Refinement | Protocol: RIGID BODY FIT | ||||||||
| Output model |  PDB-6mwq: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)