[English] 日本語
 Yorodumi
Yorodumi- EMDB-4384: Cryo-EM structure of respiratory complex I from Yarrowia lipolytica -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4384 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of respiratory complex I from Yarrowia lipolytica | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationlipoate biosynthetic process /  NADH dehydrogenase / TIM23 mitochondrial import inner membrane translocase complex / mitochondrial iron-sulfur cluster assembly complex / protein import into mitochondrial matrix / ubiquinone-6 biosynthetic process / NADH dehydrogenase / TIM23 mitochondrial import inner membrane translocase complex / mitochondrial iron-sulfur cluster assembly complex / protein import into mitochondrial matrix / ubiquinone-6 biosynthetic process /  NADH dehydrogenase activity / respiratory chain complex I / NADH dehydrogenase activity / respiratory chain complex I /  ubiquinone binding / oxidoreductase activity, acting on NAD(P)H ...lipoate biosynthetic process / ubiquinone binding / oxidoreductase activity, acting on NAD(P)H ...lipoate biosynthetic process /  NADH dehydrogenase / TIM23 mitochondrial import inner membrane translocase complex / mitochondrial iron-sulfur cluster assembly complex / protein import into mitochondrial matrix / ubiquinone-6 biosynthetic process / NADH dehydrogenase / TIM23 mitochondrial import inner membrane translocase complex / mitochondrial iron-sulfur cluster assembly complex / protein import into mitochondrial matrix / ubiquinone-6 biosynthetic process /  NADH dehydrogenase activity / respiratory chain complex I / NADH dehydrogenase activity / respiratory chain complex I /  ubiquinone binding / oxidoreductase activity, acting on NAD(P)H / ubiquinone binding / oxidoreductase activity, acting on NAD(P)H /  acyl binding / electron transport coupled proton transport / acyl binding / electron transport coupled proton transport /  iron-sulfur cluster assembly / acyl carrier activity / iron-sulfur cluster assembly / acyl carrier activity /  NADH:ubiquinone reductase (H+-translocating) / mitochondrial respiratory chain complex I / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / NADH:ubiquinone reductase (H+-translocating) / mitochondrial respiratory chain complex I / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone /  electron transport chain / electron transport chain /  NADH dehydrogenase (ubiquinone) activity / NADH dehydrogenase (ubiquinone) activity /  quinone binding / ATP synthesis coupled electron transport / quinone binding / ATP synthesis coupled electron transport /  respirasome / respirasome /  aerobic respiration / respiratory electron transport chain / aerobic respiration / respiratory electron transport chain /  mitochondrial membrane / mitochondrial membrane /  mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / fatty acid biosynthetic process / NAD binding / FMN binding / 4 iron, 4 sulfur cluster binding / response to oxidative stress / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / fatty acid biosynthetic process / NAD binding / FMN binding / 4 iron, 4 sulfur cluster binding / response to oxidative stress /  mitochondrial inner membrane / mitochondrial inner membrane /  oxidoreductase activity / protein-containing complex binding / oxidoreductase activity / protein-containing complex binding /  mitochondrion / mitochondrion /  membrane / membrane /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||
| Biological species |   Yarrowia lipolytica (yeast) Yarrowia lipolytica (yeast) | |||||||||
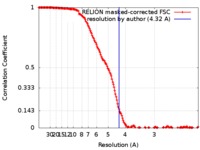
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.32 Å cryo EM / Resolution: 4.32 Å | |||||||||
 Authors Authors | Parey K / Vonck J | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: Cryo-EM structure of respiratory complex I at work. Authors: Kristian Parey / Ulrich Brandt / Hao Xie / Deryck J Mills / Karin Siegmund / Janet Vonck / Werner Kühlbrandt / Volker Zickermann /   Abstract: Mitochondrial complex I has a key role in cellular energy metabolism, generating a major portion of the proton motive force that drives aerobic ATP synthesis. The hydrophilic arm of the L-shaped ~1 ...Mitochondrial complex I has a key role in cellular energy metabolism, generating a major portion of the proton motive force that drives aerobic ATP synthesis. The hydrophilic arm of the L-shaped ~1 MDa membrane protein complex transfers electrons from NADH to ubiquinone, providing the energy to drive proton pumping at distant sites in the membrane arm. The critical steps of energy conversion are associated with the redox chemistry of ubiquinone. We report the cryo-EM structure of complete mitochondrial complex I from the aerobic yeast both in the deactive form and after capturing the enzyme during steady-state activity. The site of ubiquinone binding observed during turnover supports a two-state stabilization change mechanism for complex I. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4384.map.gz emd_4384.map.gz | 334.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4384-v30.xml emd-4384-v30.xml emd-4384.xml emd-4384.xml | 58.9 KB 58.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4384_fsc.xml emd_4384_fsc.xml | 16.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_4384.png emd_4384.png | 173.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4384 http://ftp.pdbj.org/pub/emdb/structures/EMD-4384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4384 | HTTPS FTP |
-Related structure data
| Related structure data |  6gcsMC  4385C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4384.map.gz / Format: CCP4 / Size: 361.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4384.map.gz / Format: CCP4 / Size: 361.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Mitochondrial NADH:ubiquinone oxidoreductase
+Supramolecule #1: Mitochondrial NADH:ubiquinone oxidoreductase
+Macromolecule #1: 75-KDA PROTEIN (NUAM)
+Macromolecule #2: 51-KDA PROTEIN (NUBM)
+Macromolecule #3: 49-KDA PROTEIN (NUCM)
+Macromolecule #4: NIMM SUBUNIT
+Macromolecule #5: NUEM SUBUNIT
+Macromolecule #6: NUFM SUBUNIT
+Macromolecule #7: 30-KDA PROTEIN (NUGM)
+Macromolecule #8: 24-KDA SUBUNIT (NUHM)
+Macromolecule #9: TYKY SUBUNIT (NUIM)
+Macromolecule #10: NUJM SUBUNIT
+Macromolecule #11: PSST SUBUNIT (NUKM)
+Macromolecule #12: ND4L SUBUNIT (NULM)
+Macromolecule #13: NUMM SUBUNIT
+Macromolecule #14: ACPM1 SUBUNIT
+Macromolecule #15: NB4M SUBUNIT
+Macromolecule #16: ACPM2 SUBUNIT
+Macromolecule #17: NI2M SUBUNIT
+Macromolecule #18: NESM SUBUNIT
+Macromolecule #19: NUPM SUBUNIT
+Macromolecule #20: NB6M SUBUNIT
+Macromolecule #21: NUXM SUBUNIT
+Macromolecule #22: NUYM SUBUNIT
+Macromolecule #23: NUZM SUBUNIT
+Macromolecule #24: NIAM SUBUNIT
+Macromolecule #25: NEBM SUBUNIT
+Macromolecule #26: NB2M SUBUNIT
+Macromolecule #27: NIDM SUBUNIT
+Macromolecule #28: NUUM SUBUNIT
+Macromolecule #29: NI8M SUBUNIT
+Macromolecule #30: NI9M SUBUNIT
+Macromolecule #31: N7BM SUBUNIT
+Macromolecule #32: UNKNOWN SUBUNIT
+Macromolecule #33: NB5M SUBUNIT
+Macromolecule #34: NUNM SUBUNIT
+Macromolecule #35: ND1 SUBUNIT (NU1M)
+Macromolecule #36: ND2 SUBUNIT (NU2M)
+Macromolecule #37: ND3 SUBUNIT (NU3M)
+Macromolecule #38: ND4 SUBUNIT (NU4M)
+Macromolecule #39: ND5 SUBUNIT (NU5M)
+Macromolecule #40: ND6 SUBUNIT (NU6M)
+Macromolecule #41: NB8M SUBUNIT
+Macromolecule #42: NIPM SUBUNIT
+Macromolecule #43: IRON/SULFUR CLUSTER
+Macromolecule #44: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #45: FLAVIN MONONUCLEOTIDE
+Macromolecule #46: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+Macromolecule #47: ZINC ION
+Macromolecule #48: S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-b...
+Macromolecule #49: CARDIOLIPIN
+Macromolecule #50: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| |||||||||||||||
| Grid | Model: C-flat-1/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 45872 / Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Cs: 2.0 mm / Nominal defocus max: -0.003 µm / Nominal defocus min: -0.0015 µm / Nominal magnification: 200000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 3110 / Average exposure time: 8.0 sec. / Average electron dose: 60.5 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-6gcs: |
 Movie
Movie Controller
Controller