eF-surf Help: Difference between revisions
No edit summary |
m WikiSysop moved page EF-surf Help to eF-surf Help: Revert to the original page name after adopted the wgCapitalLinks was changed to false. |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{| style="left" | |||
|__TOC__ | |||
|} | |||
== How to use == | == How to use == | ||
'''STEP-1: Up load a PDB format file.''' | '''STEP-1: Up load a PDB format file.''' | ||
| Line 45: | Line 48: | ||
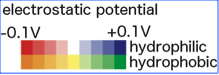
You can download efvet file and efvet-XML file with two different color schemes, one is the default color scheme (shown below as a color bar) and the other for the coloring according only to the electrostatic potential. | You can download efvet file and efvet-XML file with two different color schemes, one is the default color scheme (shown below as a color bar) and the other for the coloring according only to the electrostatic potential. | ||
[[File: | [[File:Color-bar.png|center]] | ||
The format of the efvet XML file is described in the [http://ef-site.hgc.jp/eF-site/schema/efvet30.xsd XML schema for efvet]. | |||
You can also download the object file that can be used with [http://www.avatar.se/molscript/ molscript]. | |||
The format of the object file in molscript is described [http://www.avatar.se/molscript/doc/object.html here]. | |||
That's it! However, if you find some difficulties, do not hesitate to contact us from [http://ef-site.hgc.jp/eF-site/servlet/Feedback here]. | |||
== FAQ == | |||
'''1. How long will it takes to finish the calculation?''' | |||
The calculation time will largely depend on the size of the protein. Usually, it will take 1-2 hours. Please note that the calculation exceed 6 hours will terminated to reduce the server load. | |||
'''2. What is required for calculation?''' | |||
eF-surf only needs a coordinate file in PDB format. | |||
'''3. A calculation error occurred. How can I fix it?''' | |||
There can be several reasons for calculation errors. The most probable one is the format error of the up-loaded file. Please check the upload file carefully. Frequently observed errors are due to the submission of sequence file instead of the coordinate file. eF-surf requires coordinates of proteins with PDB format. | |||
Another possibility is coming from the the size of the protein. When the calculation time exceed twelve hours, the calculation will be terminated with an error. Proteins with 500 or less residues will not encount this problem. | |||
The most difficult but less probable problem is due to unusual amino-acids in your protein. For the calculation of electrostatic potentials, we should assign the charge of every atom. Usually, the assignments of charge are done using AMBER force field, so the atoms that are not in the AMBER force field cannot be handled by eF-surf. | |||
'''4. Can the user store the calculation result more than 7 days?''' | |||
No. The limitation of 7 days to store the calculation result is due to the limited disk space in our server. | |||
'''5. How do you calculate the electrostatic potential?''' | |||
eF-surf calculate the electrostatic potential using program SCB (Nakamura & Nishida, 1987). The program solves the Poisson-Bolzmann equation numerically with a precise continuum model. | |||
'''6. What units do you use for the given electrostatic potentials?''' | |||
The unit of the electrostatic potentials is VOLT. | |||
'''7. How do you calculate the hydrophobicity?''' | |||
eF-surf use the haydrophacy indexes by Kyte and Dolittle. For every vertex on the molecular surface, the index of the nearest residues is assigned. | |||
Latest revision as of 16:29, 10 April 2018
How to use
STEP-1: Up load a PDB format file.
- Upload a PDB format file
- Input your e-mail address
- Optionally, you can give a title to your calculation (free format).
And press the Submit button. Then you will see a web page as follows;
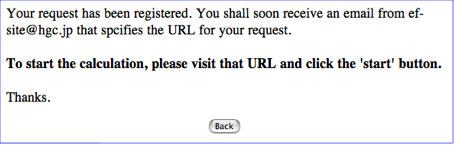
STEP-2: Check an e-mail and access to the Job-start page
You will receive an e-mail like a ...
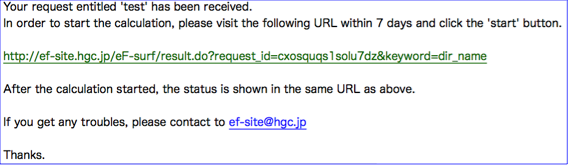
Please follow the link appeared in the e-mail ,then you will see the Job-start page as follows.
STEP-3: Start the calculation
As in the example of the Job-start page below, you will find a start button and a cancel button.
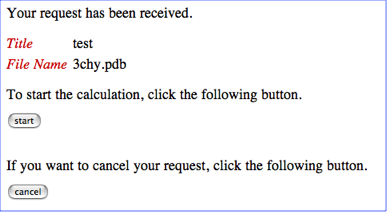
If the uploaded file name and title of your job is correct, please push the start button. Otherwise, please push the cancel button to cancel your job submission. If you push the start button, you will see the following Job-control page.
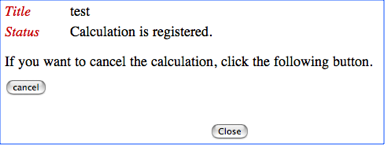
In this page you can see the status of the calculation.
STEP-4: Access to the calculation result
When the calculation is finished, you will receive another e-mail like a ...
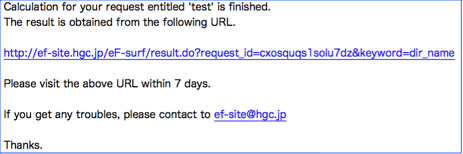
Please follow the link in the e-mail, you will reach to the result page as follows.
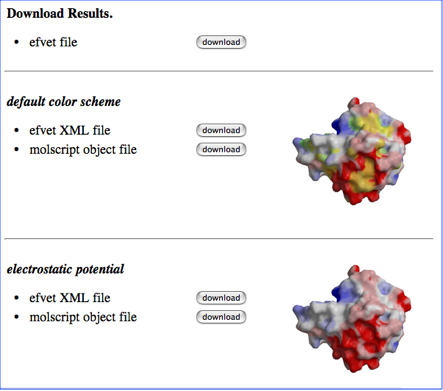
You can download efvet file and efvet-XML file with two different color schemes, one is the default color scheme (shown below as a color bar) and the other for the coloring according only to the electrostatic potential.
The format of the efvet XML file is described in the XML schema for efvet. You can also download the object file that can be used with molscript. The format of the object file in molscript is described here.
That's it! However, if you find some difficulties, do not hesitate to contact us from here.
FAQ
1. How long will it takes to finish the calculation?
The calculation time will largely depend on the size of the protein. Usually, it will take 1-2 hours. Please note that the calculation exceed 6 hours will terminated to reduce the server load.
2. What is required for calculation?
eF-surf only needs a coordinate file in PDB format.
3. A calculation error occurred. How can I fix it?
There can be several reasons for calculation errors. The most probable one is the format error of the up-loaded file. Please check the upload file carefully. Frequently observed errors are due to the submission of sequence file instead of the coordinate file. eF-surf requires coordinates of proteins with PDB format.
Another possibility is coming from the the size of the protein. When the calculation time exceed twelve hours, the calculation will be terminated with an error. Proteins with 500 or less residues will not encount this problem.
The most difficult but less probable problem is due to unusual amino-acids in your protein. For the calculation of electrostatic potentials, we should assign the charge of every atom. Usually, the assignments of charge are done using AMBER force field, so the atoms that are not in the AMBER force field cannot be handled by eF-surf.
4. Can the user store the calculation result more than 7 days?
No. The limitation of 7 days to store the calculation result is due to the limited disk space in our server.
5. How do you calculate the electrostatic potential?
eF-surf calculate the electrostatic potential using program SCB (Nakamura & Nishida, 1987). The program solves the Poisson-Bolzmann equation numerically with a precise continuum model.
6. What units do you use for the given electrostatic potentials?
The unit of the electrostatic potentials is VOLT.
7. How do you calculate the hydrophobicity?
eF-surf use the haydrophacy indexes by Kyte and Dolittle. For every vertex on the molecular surface, the index of the nearest residues is assigned.