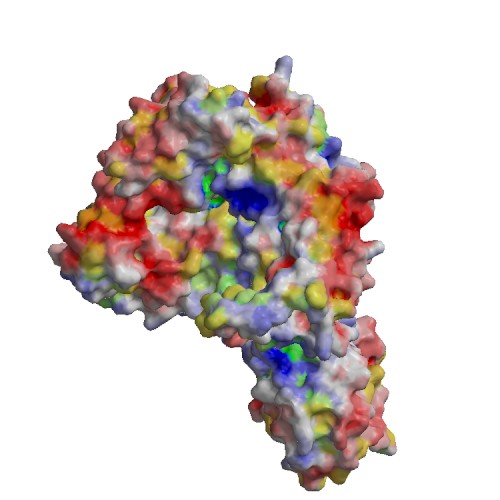
Functional site
| 1) | chain | P |
| residue | 151 | |
| type | ||
| sequence | G | |
| description | binding site for Di-peptide PLP M 801 and LYS M 367 | |
| source | : AD9 | |
| 2) | chain | P |
| residue | 398 | |
| type | ||
| sequence | T | |
| description | binding site for Di-peptide PLP M 801 and LYS M 367 | |
| source | : AD9 | |
| 3) | chain | P |
| residue | 399 | |
| type | ||
| sequence | T | |
| description | binding site for Di-peptide PLP M 801 and LYS M 367 | |
| source | : AD9 | |
| 4) | chain | P |
| residue | 400 | |
| type | ||
| sequence | S | |
| description | binding site for Di-peptide PLP M 801 and LYS M 367 | |
| source | : AD9 | |
| 5) | chain | P |
| residue | 219 | |
| type | ||
| sequence | G | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 6) | chain | P |
| residue | 220 | |
| type | ||
| sequence | T | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 7) | chain | P |
| residue | 221 | |
| type | ||
| sequence | S | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 8) | chain | P |
| residue | 245 | |
| type | ||
| sequence | H | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 9) | chain | P |
| residue | 330 | |
| type | ||
| sequence | D | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 10) | chain | P |
| residue | 332 | |
| type | ||
| sequence | A | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 11) | chain | P |
| residue | 333 | |
| type | ||
| sequence | W | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 12) | chain | P |
| residue | 364 | |
| type | ||
| sequence | S | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 13) | chain | P |
| residue | 365 | |
| type | ||
| sequence | T | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 14) | chain | P |
| residue | 366 | |
| type | ||
| sequence | H | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 15) | chain | P |
| residue | 368 | |
| type | ||
| sequence | L | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 16) | chain | P |
| residue | 369 | |
| type | ||
| sequence | L | |
| description | binding site for Di-peptide PLP P 801 and LYS P 367 | |
| source | : AE3 | |
| 17) | chain | P |
| residue | 367 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-(pyridoxal phosphate)lysine | |
| source | Swiss-Prot : SWS_FT_FI1 | |