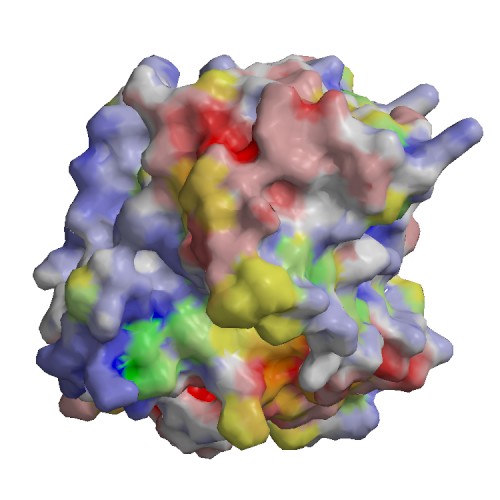
Functional site
| 1) | chain | B |
| residue | 309 | |
| type | ||
| sequence | L | |
| description | binding site for residue PFB A 404 | |
| source | : AC4 | |
| 2) | chain | B |
| residue | 218 | |
| type | ||
| sequence | R | |
| description | binding site for residue MRD A 406 | |
| source | : AC6 | |
| 3) | chain | B |
| residue | 238 | |
| type | ||
| sequence | T | |
| description | binding site for residue MRD A 406 | |
| source | : AC6 | |
| 4) | chain | B |
| residue | 239 | |
| type | ||
| sequence | E | |
| description | binding site for residue MRD A 406 | |
| source | : AC6 | |
| 5) | chain | B |
| residue | 46 | |
| type | ||
| sequence | C | |
| description | binding site for residue ZN B 401 | |
| source | : AC8 | |
| 6) | chain | B |
| residue | 67 | |
| type | ||
| sequence | H | |
| description | binding site for residue ZN B 401 | |
| source | : AC8 | |
| 7) | chain | B |
| residue | 174 | |
| type | ||
| sequence | C | |
| description | binding site for residue ZN B 401 | |
| source | : AC8 | |
| 8) | chain | B |
| residue | 97 | |
| type | ||
| sequence | C | |
| description | binding site for residue ZN B 402 | |
| source | : AC9 | |
| 9) | chain | B |
| residue | 100 | |
| type | ||
| sequence | C | |
| description | binding site for residue ZN B 402 | |
| source | : AC9 | |
| 10) | chain | B |
| residue | 103 | |
| type | ||
| sequence | C | |
| description | binding site for residue ZN B 402 | |
| source | : AC9 | |
| 11) | chain | B |
| residue | 111 | |
| type | ||
| sequence | C | |
| description | binding site for residue ZN B 402 | |
| source | : AC9 | |
| 12) | chain | B |
| residue | 47 | |
| type | ||
| sequence | R | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 13) | chain | B |
| residue | 48 | |
| type | ||
| sequence | S | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 14) | chain | B |
| residue | 51 | |
| type | ||
| sequence | H | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 15) | chain | B |
| residue | 174 | |
| type | ||
| sequence | C | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 16) | chain | B |
| residue | 178 | |
| type | ||
| sequence | T | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 17) | chain | B |
| residue | 199 | |
| type | ||
| sequence | G | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 18) | chain | B |
| residue | 201 | |
| type | ||
| sequence | G | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 19) | chain | B |
| residue | 202 | |
| type | ||
| sequence | G | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 20) | chain | B |
| residue | 203 | |
| type | ||
| sequence | V | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 21) | chain | B |
| residue | 223 | |
| type | ||
| sequence | D | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 22) | chain | B |
| residue | 224 | |
| type | ||
| sequence | I | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 23) | chain | B |
| residue | 228 | |
| type | ||
| sequence | K | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 24) | chain | B |
| residue | 268 | |
| type | ||
| sequence | V | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 25) | chain | B |
| residue | 269 | |
| type | ||
| sequence | I | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 26) | chain | B |
| residue | 292 | |
| type | ||
| sequence | V | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 27) | chain | B |
| residue | 293 | |
| type | ||
| sequence | G | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 28) | chain | B |
| residue | 294 | |
| type | ||
| sequence | V | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 29) | chain | B |
| residue | 317 | |
| type | ||
| sequence | A | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 30) | chain | B |
| residue | 318 | |
| type | ||
| sequence | I | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 31) | chain | B |
| residue | 319 | |
| type | ||
| sequence | F | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 32) | chain | B |
| residue | 369 | |
| type | ||
| sequence | R | |
| description | binding site for residue NAJ B 403 | |
| source | : AD1 | |
| 33) | chain | B |
| residue | 46 | |
| type | ||
| sequence | C | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 34) | chain | B |
| residue | 48 | |
| type | ||
| sequence | S | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 35) | chain | B |
| residue | 57 | |
| type | ||
| sequence | F | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 36) | chain | B |
| residue | 67 | |
| type | ||
| sequence | H | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 37) | chain | B |
| residue | 93 | |
| type | ||
| sequence | F | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 38) | chain | B |
| residue | 116 | |
| type | ||
| sequence | L | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 39) | chain | B |
| residue | 140 | |
| type | ||
| sequence | F | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 40) | chain | B |
| residue | 141 | |
| type | ||
| sequence | L | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 41) | chain | B |
| residue | 174 | |
| type | ||
| sequence | C | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 42) | chain | B |
| residue | 294 | |
| type | ||
| sequence | V | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 43) | chain | B |
| residue | 318 | |
| type | ||
| sequence | I | |
| description | binding site for residue PFB B 404 | |
| source | : AD2 | |
| 44) | chain | B |
| residue | 338 | |
| type | ||
| sequence | K | |
| description | binding site for residue MRD B 405 | |
| source | : AD3 | |
| 45) | chain | B |
| residue | 339 | |
| type | ||
| sequence | K | |
| description | binding site for residue MRD B 405 | |
| source | : AD3 | |
| 46) | chain | B |
| residue | 168 | |
| type | ||
| sequence | K | |
| description | binding site for residue MRD B 406 | |
| source | : AD4 | |
| 47) | chain | B |
| residue | 343 | |
| type | ||
| sequence | D | |
| description | binding site for residue MRD B 406 | |
| source | : AD4 | |
| 48) | chain | B |
| residue | 346 | |
| type | ||
| sequence | I | |
| description | binding site for residue MRD B 406 | |
| source | : AD4 | |
| 49) | chain | B |
| residue | 347 | |
| type | ||
| sequence | T | |
| description | binding site for residue MRD B 406 | |
| source | : AD4 | |
| 50) | chain | B |
| residue | 46 | |
| type | catalytic | |
| sequence | C | |
| description | 256 | |
| source | MCSA : MCSA2 | |
| 51) | chain | B |
| residue | 48 | |
| type | catalytic | |
| sequence | S | |
| description | 256 | |
| source | MCSA : MCSA2 | |
| 52) | chain | B |
| residue | 51 | |
| type | catalytic | |
| sequence | H | |
| description | 256 | |
| source | MCSA : MCSA2 | |
| 53) | chain | B |
| residue | 67 | |
| type | catalytic | |
| sequence | H | |
| description | 256 | |
| source | MCSA : MCSA2 | |
| 54) | chain | B |
| residue | 174 | |
| type | catalytic | |
| sequence | C | |
| description | 256 | |
| source | MCSA : MCSA2 | |
| 55) | chain | B |
| residue | 174 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1A72, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1ADC, ECO:0007744|PDB:1ADF, ECO:0007744|PDB:1ADG, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QLH, ECO:0007744|PDB:1QLJ, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:1YE3, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:5ADH, ECO:0007744|PDB:6ADH, ECO:0007744|PDB:7ADH, ECO:0007744|PDB:8ADH | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 56) | chain | B |
| residue | 48 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000250|UniProtKB:P06525 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 57) | chain | B |
| residue | 292 | |
| type | BINDING | |
| sequence | V | |
| description | BINDING => ECO:0000250|UniProtKB:P06525 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 58) | chain | B |
| residue | 319 | |
| type | BINDING | |
| sequence | F | |
| description | BINDING => ECO:0000250|UniProtKB:P06525 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 59) | chain | B |
| residue | 1 | |
| type | MOD_RES | |
| sequence | S | |
| description | N-acetylserine => ECO:0000269|PubMed:5466062 | |
| source | Swiss-Prot : SWS_FT_FI9 | |
| 60) | chain | B |
| residue | 46 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0000269|PubMed:178875, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1A72, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1ADC, ECO:0007744|PDB:1ADF, ECO:0007744|PDB:1ADG, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QLH, ECO:0007744|PDB:1QLJ, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:1YE3, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:5ADH, ECO:0007744|PDB:6ADH, ECO:0007744|PDB:7ADH, ECO:0007744|PDB:8ADH | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 61) | chain | B |
| residue | 67 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0000269|PubMed:178875, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1A72, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1ADC, ECO:0007744|PDB:1ADF, ECO:0007744|PDB:1ADG, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QLH, ECO:0007744|PDB:1QLJ, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:1YE3, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:5ADH, ECO:0007744|PDB:6ADH, ECO:0007744|PDB:7ADH, ECO:0007744|PDB:8ADH | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 62) | chain | B |
| residue | 97 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0000269|PubMed:178875, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1A72, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1ADC, ECO:0007744|PDB:1ADF, ECO:0007744|PDB:1ADG, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QLH, ECO:0007744|PDB:1QLJ, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:1YE3, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:2OXI, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:5ADH, ECO:0007744|PDB:6ADH, ECO:0007744|PDB:7ADH, ECO:0007744|PDB:8ADH | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 63) | chain | B |
| residue | 100 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0000269|PubMed:178875, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1A72, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1ADC, ECO:0007744|PDB:1ADF, ECO:0007744|PDB:1ADG, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QLH, ECO:0007744|PDB:1QLJ, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:1YE3, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:2OXI, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:5ADH, ECO:0007744|PDB:6ADH, ECO:0007744|PDB:7ADH, ECO:0007744|PDB:8ADH | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 64) | chain | B |
| residue | 103 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0000269|PubMed:178875, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1A72, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1ADC, ECO:0007744|PDB:1ADF, ECO:0007744|PDB:1ADG, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QLH, ECO:0007744|PDB:1QLJ, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:1YE3, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:2OXI, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:5ADH, ECO:0007744|PDB:6ADH, ECO:0007744|PDB:7ADH, ECO:0007744|PDB:8ADH | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 65) | chain | B |
| residue | 111 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0000269|PubMed:178875, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1A72, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1ADC, ECO:0007744|PDB:1ADF, ECO:0007744|PDB:1ADG, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QLH, ECO:0007744|PDB:1QLJ, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:1YE3, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:2OXI, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:5ADH, ECO:0007744|PDB:6ADH, ECO:0007744|PDB:7ADH, ECO:0007744|PDB:8ADH | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 66) | chain | B |
| residue | 199 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HEU, ECO:0007744|PDB:1HF3, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:2JHF, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:2OXI, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:6ADH | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 67) | chain | B |
| residue | 223 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HEU, ECO:0007744|PDB:1HF3, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QLH, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:2JHF, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:2OXI, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:5ADH, ECO:0007744|PDB:6ADH | |
| source | Swiss-Prot : SWS_FT_FI6 | |
| 68) | chain | B |
| residue | 228 | |
| type | BINDING | |
| sequence | K | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HEU, ECO:0007744|PDB:1HF3, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1JU9, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1QLH, ECO:0007744|PDB:2JHF, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:2OXI, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:5ADH, ECO:0007744|PDB:6ADH | |
| source | Swiss-Prot : SWS_FT_FI7 | |
| 69) | chain | B |
| residue | 369 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000269|PubMed:15299346, ECO:0007744|PDB:1A71, ECO:0007744|PDB:1ADB, ECO:0007744|PDB:1AXE, ECO:0007744|PDB:1AXG, ECO:0007744|PDB:1BTO, ECO:0007744|PDB:1HET, ECO:0007744|PDB:1HEU, ECO:0007744|PDB:1HF3, ECO:0007744|PDB:1HLD, ECO:0007744|PDB:1LDE, ECO:0007744|PDB:1LDY, ECO:0007744|PDB:1MG0, ECO:0007744|PDB:1MGO, ECO:0007744|PDB:1N8K, ECO:0007744|PDB:1N92, ECO:0007744|PDB:1P1R, ECO:0007744|PDB:1QV6, ECO:0007744|PDB:1QV7, ECO:0007744|PDB:2JHF, ECO:0007744|PDB:2JHG, ECO:0007744|PDB:2OHX, ECO:0007744|PDB:2OXI, ECO:0007744|PDB:3BTO, ECO:0007744|PDB:3OQ6, ECO:0007744|PDB:4DWV, ECO:0007744|PDB:4DXH, ECO:0007744|PDB:4NFH, ECO:0007744|PDB:4NFS, ECO:0007744|PDB:4NG5, ECO:0007744|PDB:4XD2, ECO:0007744|PDB:6ADH | |
| source | Swiss-Prot : SWS_FT_FI8 | |