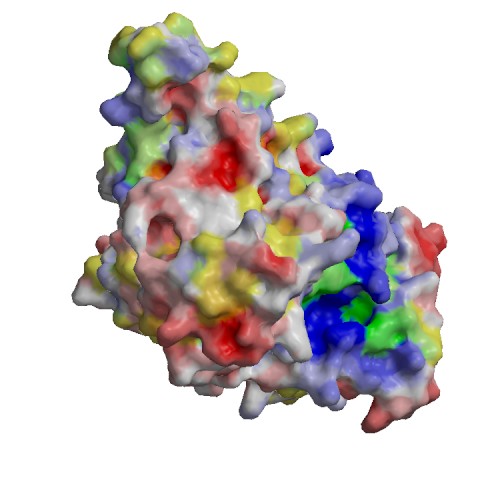
Functional site
| 1) | chain | B |
| residue | 497 | |
| type | ||
| sequence | L | |
| description | binding site for residue CPS A 605 | |
| source | : AC5 | |
| 2) | chain | B |
| residue | 529 | |
| type | ||
| sequence | Y | |
| description | binding site for residue CPS A 605 | |
| source | : AC5 | |
| 3) | chain | B |
| residue | 532 | |
| type | ||
| sequence | Y | |
| description | binding site for residue CPS A 605 | |
| source | : AC5 | |
| 4) | chain | B |
| residue | 515 | |
| type | ||
| sequence | N | |
| description | binding site for residue CPS A 606 | |
| source | : AC6 | |
| 5) | chain | B |
| residue | 525 | |
| type | ||
| sequence | A | |
| description | binding site for residue CPS A 606 | |
| source | : AC6 | |
| 6) | chain | B |
| residue | 551 | |
| type | ||
| sequence | G | |
| description | binding site for residue CPS A 608 | |
| source | : AC8 | |
| 7) | chain | B |
| residue | 456 | |
| type | ||
| sequence | W | |
| description | binding site for residue CPS A 609 | |
| source | : AC9 | |
| 8) | chain | B |
| residue | 213 | |
| type | ||
| sequence | S | |
| description | binding site for residue GOL B 601 | |
| source | : AD3 | |
| 9) | chain | B |
| residue | 215 | |
| type | ||
| sequence | N | |
| description | binding site for residue GOL B 601 | |
| source | : AD3 | |
| 10) | chain | B |
| residue | 219 | |
| type | ||
| sequence | Q | |
| description | binding site for residue GOL B 601 | |
| source | : AD3 | |
| 11) | chain | B |
| residue | 310 | |
| type | ||
| sequence | L | |
| description | binding site for residue SO4 B 602 | |
| source | : AD4 | |
| 12) | chain | B |
| residue | 343 | |
| type | ||
| sequence | R | |
| description | binding site for residue SO4 B 602 | |
| source | : AD4 | |
| 13) | chain | B |
| residue | 386 | |
| type | ||
| sequence | R | |
| description | binding site for residue SO4 B 602 | |
| source | : AD4 | |
| 14) | chain | B |
| residue | 555 | |
| type | ||
| sequence | Y | |
| description | binding site for residue CPS B 603 | |
| source | : AD5 | |
| 15) | chain | B |
| residue | 559 | |
| type | ||
| sequence | S | |
| description | binding site for residue CPS B 603 | |
| source | : AD5 | |
| 16) | chain | B |
| residue | 562 | |
| type | ||
| sequence | F | |
| description | binding site for residue CPS B 604 | |
| source | : AD6 | |
| 17) | chain | B |
| residue | 566 | |
| type | ||
| sequence | Y | |
| description | binding site for residue CPS B 604 | |
| source | : AD6 | |
| 18) | chain | B |
| residue | 166 | |
| type | ||
| sequence | F | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 19) | chain | B |
| residue | 195 | |
| type | ||
| sequence | Y | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 20) | chain | B |
| residue | 210 | |
| type | ||
| sequence | Y | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 21) | chain | B |
| residue | 322 | |
| type | ||
| sequence | A | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 22) | chain | B |
| residue | 333 | |
| type | ||
| sequence | L | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 23) | chain | B |
| residue | 335 | |
| type | ||
| sequence | Y | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 24) | chain | B |
| residue | 415 | |
| type | ||
| sequence | P | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 25) | chain | B |
| residue | 416 | |
| type | ||
| sequence | L | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 26) | chain | B |
| residue | 418 | |
| type | ||
| sequence | G | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 27) | chain | B |
| residue | 473 | |
| type | ||
| sequence | L | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 28) | chain | B |
| residue | 477 | |
| type | ||
| sequence | F | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 29) | chain | B |
| residue | 495 | |
| type | ||
| sequence | F | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 30) | chain | B |
| residue | 505 | |
| type | ||
| sequence | P | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 31) | chain | B |
| residue | 506 | |
| type | ||
| sequence | V | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 32) | chain | B |
| residue | 509 | |
| type | ||
| sequence | L | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 33) | chain | B |
| residue | 519 | |
| type | ||
| sequence | L | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 34) | chain | B |
| residue | 522 | |
| type | ||
| sequence | H | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 35) | chain | B |
| residue | 523 | |
| type | ||
| sequence | F | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 36) | chain | B |
| residue | 526 | |
| type | ||
| sequence | V | |
| description | binding site for residue EKV B 605 | |
| source | : AD7 | |
| 37) | chain | B |
| residue | 130 | |
| type | ||
| sequence | G | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 38) | chain | B |
| residue | 132 | |
| type | ||
| sequence | G | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 39) | chain | B |
| residue | 133 | |
| type | ||
| sequence | V | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 40) | chain | B |
| residue | 134 | |
| type | ||
| sequence | L | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 41) | chain | B |
| residue | 152 | |
| type | ||
| sequence | I | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 42) | chain | B |
| residue | 153 | |
| type | ||
| sequence | E | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 43) | chain | B |
| residue | 154 | |
| type | ||
| sequence | R | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 44) | chain | B |
| residue | 161 | |
| type | ||
| sequence | R | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 45) | chain | B |
| residue | 163 | |
| type | ||
| sequence | V | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 46) | chain | B |
| residue | 164 | |
| type | ||
| sequence | G | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 47) | chain | B |
| residue | 165 | |
| type | ||
| sequence | E | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 48) | chain | B |
| residue | 166 | |
| type | ||
| sequence | F | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 49) | chain | B |
| residue | 234 | |
| type | ||
| sequence | R | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 50) | chain | B |
| residue | 250 | |
| type | ||
| sequence | V | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 51) | chain | B |
| residue | 285 | |
| type | ||
| sequence | D | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 52) | chain | B |
| residue | 286 | |
| type | ||
| sequence | G | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 53) | chain | B |
| residue | 389 | |
| type | ||
| sequence | P | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 54) | chain | B |
| residue | 407 | |
| type | ||
| sequence | G | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 55) | chain | B |
| residue | 408 | |
| type | ||
| sequence | D | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 56) | chain | B |
| residue | 415 | |
| type | ||
| sequence | P | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 57) | chain | B |
| residue | 418 | |
| type | ||
| sequence | G | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 58) | chain | B |
| residue | 420 | |
| type | ||
| sequence | G | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 59) | chain | B |
| residue | 421 | |
| type | ||
| sequence | M | |
| description | binding site for residue FAD B 606 | |
| source | : AD8 | |
| 60) | chain | B |
| residue | 153 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000269|PubMed:30626872, ECO:0007744|PDB:6C6N | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 61) | chain | B |
| residue | 161 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000269|PubMed:30626872, ECO:0007744|PDB:6C6N | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 62) | chain | B |
| residue | 166 | |
| type | BINDING | |
| sequence | F | |
| description | BINDING => ECO:0000269|PubMed:30626872, ECO:0007744|PDB:6C6N | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 63) | chain | B |
| residue | 234 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000269|PubMed:30626872, ECO:0007744|PDB:6C6N | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 64) | chain | B |
| residue | 250 | |
| type | BINDING | |
| sequence | V | |
| description | BINDING => ECO:0000269|PubMed:30626872, ECO:0007744|PDB:6C6N | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 65) | chain | B |
| residue | 408 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:30626872, ECO:0007744|PDB:6C6N | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 66) | chain | B |
| residue | 421 | |
| type | BINDING | |
| sequence | M | |
| description | BINDING => ECO:0000269|PubMed:30626872, ECO:0007744|PDB:6C6N | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 67) | chain | B |
| residue | 133 | |
| type | BINDING | |
| sequence | V | |
| description | BINDING => ECO:0000269|PubMed:30626872, ECO:0007744|PDB:6C6N | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 68) | chain | B |
| residue | 195 | |
| type | SITE | |
| sequence | Y | |
| description | Important for enzyme activity => ECO:0000269|PubMed:30626872 | |
| source | Swiss-Prot : SWS_FT_FI2 | |