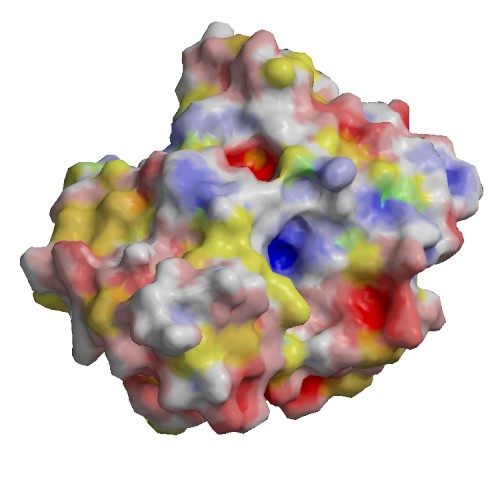
Functional site
| 1) | chain | D |
| residue | 109 | |
| type | ||
| sequence | Q | |
| description | binding site for residue PO4 C 301 | |
| source | : AC5 | |
| 2) | chain | D |
| residue | 217 | |
| type | ||
| sequence | T | |
| description | binding site for residue 9F2 C 302 | |
| source | : AC6 | |
| 3) | chain | D |
| residue | 218 | |
| type | ||
| sequence | T | |
| description | binding site for residue 9F2 C 302 | |
| source | : AC6 | |
| 4) | chain | D |
| residue | 241 | |
| type | ||
| sequence | D | |
| description | binding site for residue 9F2 C 302 | |
| source | : AC6 | |
| 5) | chain | D |
| residue | 70 | |
| type | ||
| sequence | S | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 6) | chain | D |
| residue | 128 | |
| type | ||
| sequence | S | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 7) | chain | D |
| residue | 169 | |
| type | ||
| sequence | P | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 8) | chain | D |
| residue | 172 | |
| type | ||
| sequence | N | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 9) | chain | D |
| residue | 173 | |
| type | ||
| sequence | R | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 10) | chain | D |
| residue | 218 | |
| type | ||
| sequence | T | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 11) | chain | D |
| residue | 236 | |
| type | ||
| sequence | K | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 12) | chain | D |
| residue | 237 | |
| type | ||
| sequence | T | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 13) | chain | D |
| residue | 238 | |
| type | ||
| sequence | G | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 14) | chain | D |
| residue | 239 | |
| type | ||
| sequence | T | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 15) | chain | D |
| residue | 240 | |
| type | ||
| sequence | G | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 16) | chain | D |
| residue | 241 | |
| type | ||
| sequence | D | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 17) | chain | D |
| residue | 278 | |
| type | ||
| sequence | E | |
| description | binding site for residue 9F2 D 400 | |
| source | : AC7 | |
| 18) | chain | D |
| residue | 70 | |
| type | ACT_SITE | |
| sequence | S | |
| description | Acyl-ester intermediate => ECO:0000269|PubMed:19251630, ECO:0000269|PubMed:20353175, ECO:0000269|PubMed:20961112 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 19) | chain | D |
| residue | 168 | |
| type | ACT_SITE | |
| sequence | E | |
| description | Proton acceptor => ECO:0000303|PubMed:20353175, ECO:0000303|PubMed:20961112 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 20) | chain | D |
| residue | 128 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:19251630, ECO:0000269|PubMed:20353175, ECO:0000269|PubMed:20961112 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 21) | chain | D |
| residue | 237 | |
| type | BINDING | |
| sequence | T | |
| description | BINDING => ECO:0000269|PubMed:19251630, ECO:0000269|PubMed:20353175, ECO:0000269|PubMed:20961112 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 22) | chain | D |
| residue | 73 | |
| type | SITE | |
| sequence | K | |
| description | Increases nucleophilicity of active site Ser => ECO:0000303|PubMed:20353175, ECO:0000303|PubMed:20961112 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 23) | chain | D |
| residue | 103 | |
| type | SITE | |
| sequence | I | |
| description | Functions as a gatekeeper residue that regulates substrate accessibility to the enzyme active site => ECO:0000303|PubMed:24023821 | |
| source | Swiss-Prot : SWS_FT_FI5 | |