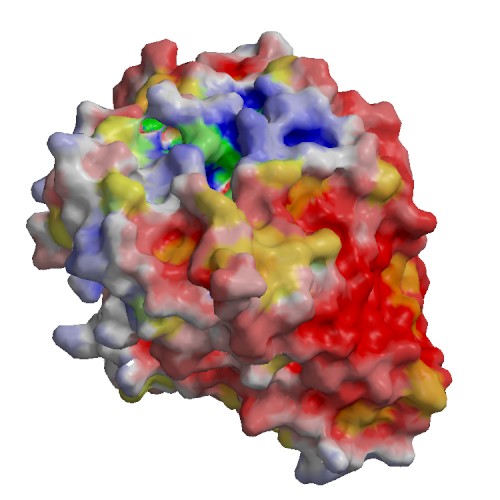
Functional site
| 1) | chain | D |
| residue | 252 | |
| type | ||
| sequence | K | |
| description | binding site for residue GTP C 501 | |
| source | : AC9 | |
| 2) | chain | D |
| residue | 10 | |
| type | ||
| sequence | G | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 3) | chain | D |
| residue | 11 | |
| type | ||
| sequence | Q | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 4) | chain | D |
| residue | 12 | |
| type | ||
| sequence | C | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 5) | chain | D |
| residue | 15 | |
| type | ||
| sequence | Q | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 6) | chain | D |
| residue | 97 | |
| type | ||
| sequence | A | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 7) | chain | D |
| residue | 98 | |
| type | ||
| sequence | G | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 8) | chain | D |
| residue | 99 | |
| type | ||
| sequence | N | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 9) | chain | D |
| residue | 138 | |
| type | ||
| sequence | S | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 10) | chain | D |
| residue | 141 | |
| type | ||
| sequence | G | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 11) | chain | D |
| residue | 142 | |
| type | ||
| sequence | G | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 12) | chain | D |
| residue | 143 | |
| type | ||
| sequence | T | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 13) | chain | D |
| residue | 144 | |
| type | ||
| sequence | G | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 14) | chain | D |
| residue | 175 | |
| type | ||
| sequence | V | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 15) | chain | D |
| residue | 181 | |
| type | ||
| sequence | E | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 16) | chain | D |
| residue | 204 | |
| type | ||
| sequence | N | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 17) | chain | D |
| residue | 222 | |
| type | ||
| sequence | Y | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 18) | chain | D |
| residue | 226 | |
| type | ||
| sequence | N | |
| description | binding site for residue GTP D 501 | |
| source | : AD3 | |
| 19) | chain | D |
| residue | 11 | |
| type | ||
| sequence | Q | |
| description | binding site for residue MG D 502 | |
| source | : AD4 | |
| 20) | chain | D |
| residue | 239 | |
| type | ||
| sequence | C | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 21) | chain | D |
| residue | 248 | |
| type | ||
| sequence | A | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 22) | chain | D |
| residue | 252 | |
| type | ||
| sequence | K | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 23) | chain | D |
| residue | 253 | |
| type | ||
| sequence | L | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 24) | chain | D |
| residue | 256 | |
| type | ||
| sequence | N | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 25) | chain | D |
| residue | 257 | |
| type | ||
| sequence | M | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 26) | chain | D |
| residue | 315 | |
| type | ||
| sequence | A | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 27) | chain | D |
| residue | 316 | |
| type | ||
| sequence | I | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 28) | chain | D |
| residue | 348 | |
| type | ||
| sequence | N | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 29) | chain | D |
| residue | 350 | |
| type | ||
| sequence | K | |
| description | binding site for residue 890 D 503 | |
| source | : AD5 | |
| 30) | chain | D |
| residue | 58 | |
| type | CROSSLNK | |
| sequence | K | |
| description | Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in ubiquitin); alternate => ECO:0000250|UniProtKB:P07437 | |
| source | Swiss-Prot : SWS_FT_FI9 | |
| 31) | chain | D |
| residue | 142 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000250|UniProtKB:Q13509 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 32) | chain | D |
| residue | 143 | |
| type | BINDING | |
| sequence | T | |
| description | BINDING => ECO:0000250|UniProtKB:Q13509 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 33) | chain | D |
| residue | 144 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000250|UniProtKB:Q13509 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 34) | chain | D |
| residue | 204 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000250|UniProtKB:Q13509 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 35) | chain | D |
| residue | 226 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000250|UniProtKB:Q13509 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 36) | chain | D |
| residue | 11 | |
| type | BINDING | |
| sequence | Q | |
| description | BINDING => ECO:0000250|UniProtKB:Q13509 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 37) | chain | D |
| residue | 138 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000250|UniProtKB:Q13509 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 38) | chain | D |
| residue | 69 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000250|UniProtKB:P68363 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 39) | chain | D |
| residue | 40 | |
| type | MOD_RES | |
| sequence | S | |
| description | Phosphoserine => ECO:0000250|UniProtKB:P99024 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 40) | chain | D |
| residue | 58 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-succinyllysine; alternate => ECO:0000250|UniProtKB:P99024 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 41) | chain | D |
| residue | 172 | |
| type | MOD_RES | |
| sequence | S | |
| description | Phosphoserine; by CDK1 => ECO:0000250|UniProtKB:Q13885 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 42) | chain | D |
| residue | 285 | |
| type | MOD_RES | |
| sequence | T | |
| description | Phosphothreonine => ECO:0000250|UniProtKB:P07437 | |
| source | Swiss-Prot : SWS_FT_FI6 | |
| 43) | chain | D |
| residue | 290 | |
| type | MOD_RES | |
| sequence | T | |
| description | Phosphothreonine => ECO:0000250|UniProtKB:P07437 | |
| source | Swiss-Prot : SWS_FT_FI6 | |
| 44) | chain | D |
| residue | 318 | |
| type | MOD_RES | |
| sequence | R | |
| description | Omega-N-methylarginine => ECO:0000250|UniProtKB:P07437 | |
| source | Swiss-Prot : SWS_FT_FI7 | |
| 45) | chain | D |
| residue | 324 | |
| type | CROSSLNK | |
| sequence | K | |
| description | Glycyl lysine isopeptide (Lys-Gly) (interchain with G-Cter in ubiquitin) => ECO:0000250|UniProtKB:P07437 | |
| source | Swiss-Prot : SWS_FT_FI10 | |