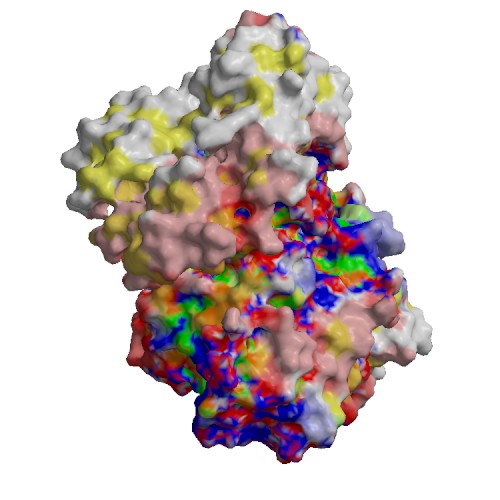
Functional site
| 1) | chain | A |
| residue | 19 | |
| type | ||
| sequence | R | |
| description | binding site for residue CL A 501 | |
| source | : AC1 | |
| 2) | chain | B |
| residue | 178 | |
| type | ||
| sequence | L | |
| description | binding site for residue CL A 501 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 297 | |
| type | ||
| sequence | T | |
| description | binding site for residue PEG A 504 | |
| source | : AC2 | |
| 4) | chain | A |
| residue | 299 | |
| type | ||
| sequence | S | |
| description | binding site for residue PEG A 504 | |
| source | : AC2 | |
| 5) | chain | A |
| residue | 301 | |
| type | ||
| sequence | E | |
| description | binding site for residue PEG A 504 | |
| source | : AC2 | |
| 6) | chain | B |
| residue | 172 | |
| type | ||
| sequence | S | |
| description | binding site for residue CL B 501 | |
| source | : AC3 | |
| 7) | chain | B |
| residue | 291 | |
| type | ||
| sequence | T | |
| description | binding site for residue CL B 501 | |
| source | : AC3 | |
| 8) | chain | B |
| residue | 292 | |
| type | ||
| sequence | C | |
| description | binding site for residue CL B 501 | |
| source | : AC3 | |
| 9) | chain | B |
| residue | 293 | |
| type | ||
| sequence | S | |
| description | binding site for residue CL B 501 | |
| source | : AC3 | |
| 10) | chain | B |
| residue | 294 | |
| type | ||
| sequence | Q | |
| description | binding site for residue CL B 501 | |
| source | : AC3 | |
| 11) | chain | B |
| residue | 342 | |
| type | ||
| sequence | N | |
| description | binding site for residue CL B 501 | |
| source | : AC3 | |
| 12) | chain | A |
| residue | 315 | |
| type | ||
| sequence | F | |
| description | binding site for residue MG B 502 | |
| source | : AC4 | |
| 13) | chain | A |
| residue | 316 | |
| type | ||
| sequence | S | |
| description | binding site for residue MG B 502 | |
| source | : AC4 | |
| 14) | chain | B |
| residue | 6 | |
| type | ||
| sequence | P | |
| description | binding site for residue MG B 502 | |
| source | : AC4 | |
| 15) | chain | B |
| residue | 8 | |
| type | ||
| sequence | W | |
| description | binding site for residue MG B 502 | |
| source | : AC4 | |
| 16) | chain | B |
| residue | 11 | |
| type | ||
| sequence | Q | |
| description | binding site for residue MG B 502 | |
| source | : AC4 | |
| 17) | chain | A |
| residue | 178 | |
| type | ||
| sequence | L | |
| description | binding site for residue IMD B 503 | |
| source | : AC5 | |
| 18) | chain | A |
| residue | 179 | |
| type | ||
| sequence | N | |
| description | binding site for residue IMD B 503 | |
| source | : AC5 | |
| 19) | chain | B |
| residue | 19 | |
| type | ||
| sequence | R | |
| description | binding site for residue IMD B 503 | |
| source | : AC5 | |
| 20) | chain | B |
| residue | 184 | |
| type | ||
| sequence | E | |
| description | binding site for residue IMD B 503 | |
| source | : AC5 | |
| 21) | chain | B |
| residue | 209 | |
| type | ||
| sequence | Y | |
| description | binding site for residue IMD B 503 | |
| source | : AC5 | |
| 22) | chain | A |
| residue | 153 | |
| type | ||
| sequence | S | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 23) | chain | A |
| residue | 196 | |
| type | ||
| sequence | H | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 24) | chain | A |
| residue | 197 | |
| type | ||
| sequence | I | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 25) | chain | A |
| residue | 198 | |
| type | ||
| sequence | S | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 26) | chain | A |
| residue | 238 | |
| type | ||
| sequence | A | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 27) | chain | A |
| residue | 261 | |
| type | ||
| sequence | L | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 28) | chain | A |
| residue | 263 | |
| type | ||
| sequence | G | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 29) | chain | A |
| residue | 264 | |
| type | ||
| sequence | G | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 30) | chain | A |
| residue | 265 | |
| type | ||
| sequence | G | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 31) | chain | A |
| residue | 266 | |
| type | ||
| sequence | P | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 32) | chain | A |
| residue | 285 | |
| type | ||
| sequence | S | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 33) | chain | A |
| residue | 287 | |
| type | ||
| sequence | G | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 34) | chain | A |
| residue | 288 | |
| type | ||
| sequence | M | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 35) | chain | A |
| residue | 289 | |
| type | ||
| sequence | T | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 36) | chain | A |
| residue | 293 | |
| type | ||
| sequence | S | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 37) | chain | A |
| residue | 294 | |
| type | ||
| sequence | Q | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 38) | chain | A |
| residue | 367 | |
| type | ||
| sequence | D | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 39) | chain | A |
| residue | 379 | |
| type | ||
| sequence | V | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 40) | chain | A |
| residue | 382 | |
| type | ||
| sequence | R | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 41) | chain | A |
| residue | 471 | |
| type | ||
| sequence | K | |
| description | binding site for Di-nucleotide A A 505 and OSB A 506 | |
| source | : AC6 | |
| 42) | chain | B |
| residue | 197 | |
| type | ||
| sequence | I | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 43) | chain | B |
| residue | 198 | |
| type | ||
| sequence | S | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 44) | chain | B |
| residue | 238 | |
| type | ||
| sequence | A | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 45) | chain | B |
| residue | 261 | |
| type | ||
| sequence | L | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 46) | chain | B |
| residue | 263 | |
| type | ||
| sequence | G | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 47) | chain | B |
| residue | 264 | |
| type | ||
| sequence | G | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 48) | chain | B |
| residue | 266 | |
| type | ||
| sequence | P | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 49) | chain | B |
| residue | 285 | |
| type | ||
| sequence | S | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 50) | chain | B |
| residue | 287 | |
| type | ||
| sequence | G | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 51) | chain | B |
| residue | 288 | |
| type | ||
| sequence | M | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 52) | chain | B |
| residue | 289 | |
| type | ||
| sequence | T | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 53) | chain | B |
| residue | 293 | |
| type | ||
| sequence | S | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 54) | chain | B |
| residue | 294 | |
| type | ||
| sequence | Q | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 55) | chain | B |
| residue | 367 | |
| type | ||
| sequence | D | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 56) | chain | B |
| residue | 379 | |
| type | ||
| sequence | V | |
| description | binding site for Di-nucleotide A B 504 and OSB B 505 | |
| source | : AC7 | |
| 57) | chain | A |
| residue | 149-160 | |
| type | prosite | |
| sequence | LMYTSGTTGKPK | |
| description | AMP_BINDING Putative AMP-binding domain signature. LMYTSGTTGkPK | |
| source | prosite : PS00455 | |