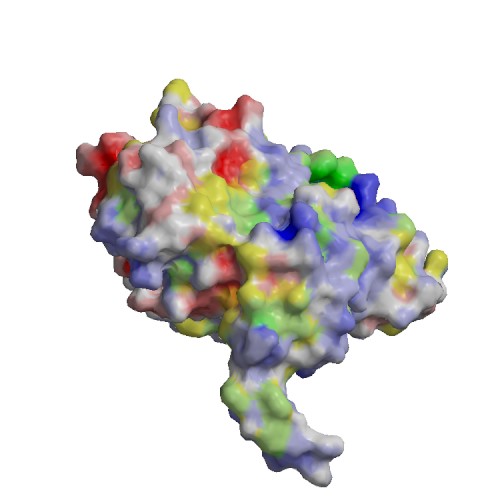
Functional site
| 1) | chain | A |
| residue | 416 | |
| type | ||
| sequence | G | |
| description | binding site for residue MG A 601 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 417 | |
| type | ||
| sequence | S | |
| description | binding site for residue MG A 601 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 427 | |
| type | ||
| sequence | D | |
| description | binding site for residue MG A 601 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 429 | |
| type | ||
| sequence | D | |
| description | binding site for residue MG A 601 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 427 | |
| type | ||
| sequence | D | |
| description | binding site for residue MN A 602 | |
| source | : AC2 | |
| 6) | chain | A |
| residue | 429 | |
| type | ||
| sequence | D | |
| description | binding site for residue MN A 602 | |
| source | : AC2 | |
| 7) | chain | A |
| residue | 490 | |
| type | ||
| sequence | D | |
| description | binding site for residue MN A 602 | |
| source | : AC2 | |
| 8) | chain | A |
| residue | 386 | |
| type | ||
| sequence | R | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 9) | chain | A |
| residue | 416 | |
| type | ||
| sequence | G | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 10) | chain | A |
| residue | 417 | |
| type | ||
| sequence | S | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 11) | chain | A |
| residue | 420 | |
| type | ||
| sequence | R | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 12) | chain | A |
| residue | 426 | |
| type | ||
| sequence | G | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 13) | chain | A |
| residue | 427 | |
| type | ||
| sequence | D | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 14) | chain | A |
| residue | 429 | |
| type | ||
| sequence | D | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 15) | chain | A |
| residue | 505 | |
| type | ||
| sequence | Y | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 16) | chain | A |
| residue | 506 | |
| type | ||
| sequence | F | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 17) | chain | A |
| residue | 507 | |
| type | ||
| sequence | T | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 18) | chain | A |
| residue | 508 | |
| type | ||
| sequence | G | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 19) | chain | A |
| residue | 513 | |
| type | ||
| sequence | N | |
| description | binding site for residue DCP A 603 | |
| source | : AC3 | |
| 20) | chain | A |
| residue | 339 | |
| type | ||
| sequence | S | |
| description | binding site for residue MN A 604 | |
| source | : AC4 | |
| 21) | chain | A |
| residue | 341 | |
| type | ||
| sequence | I | |
| description | binding site for residue MN A 604 | |
| source | : AC4 | |
| 22) | chain | A |
| residue | 344 | |
| type | ||
| sequence | A | |
| description | binding site for residue MN A 604 | |
| source | : AC4 | |
| 23) | chain | A |
| residue | 312 | |
| type | ACT_SITE | |
| sequence | K | |
| description | Schiff-base intermediate with DNA => ECO:0000269|PubMed:11457865 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 24) | chain | A |
| residue | 386 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000269|PubMed:17475573, ECO:0007744|PDB:2PFP, ECO:0007744|PDB:2PFQ | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 25) | chain | A |
| residue | 417 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:17475573, ECO:0007744|PDB:2PFP, ECO:0007744|PDB:2PFQ | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 26) | chain | A |
| residue | 426 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000269|PubMed:17475573, ECO:0007744|PDB:2PFP, ECO:0007744|PDB:2PFQ | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 27) | chain | A |
| residue | 513 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000269|PubMed:17475573, ECO:0007744|PDB:2PFP, ECO:0007744|PDB:2PFQ | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 28) | chain | A |
| residue | 427 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:17475573, ECO:0007744|PDB:2PFO, ECO:0007744|PDB:2PFQ | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 29) | chain | A |
| residue | 429 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:17475573, ECO:0007744|PDB:2PFO, ECO:0007744|PDB:2PFQ | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 30) | chain | A |
| residue | 490 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:17475573, ECO:0007744|PDB:2PFO, ECO:0007744|PDB:2PFQ | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 31) | chain | A |
| residue | 416-435 | |
| type | prosite | |
| sequence | GSYRRGKATCGDVDVLITHP | |
| description | DNA_POLYMERASE_X DNA polymerase family X signature. GSYrRGkatCgDVDVLIthP | |
| source | prosite : PS00522 | |