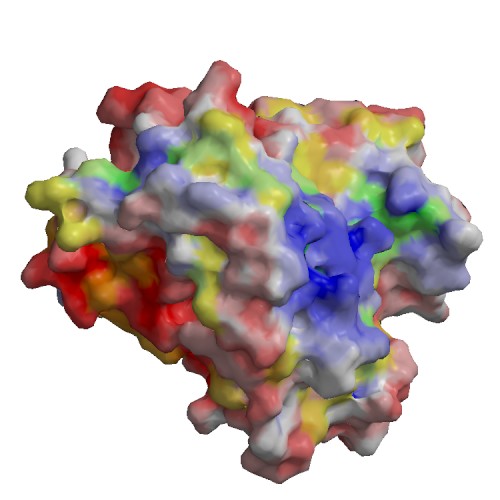
Functional site
| 1) | chain | A |
| residue | 60 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 61 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 64 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 68 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 86 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 6) | chain | A |
| residue | 89 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 7) | chain | A |
| residue | 91 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 8) | chain | A |
| residue | 125 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 9) | chain | A |
| residue | 140 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 10) | chain | A |
| residue | 142 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 11) | chain | B |
| residue | 60 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE FSE A 201 | |
| source | : AC1 | |
| 12) | chain | A |
| residue | 55 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE GOL A 202 | |
| source | : AC2 | |
| 13) | chain | A |
| residue | 56 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE GOL A 202 | |
| source | : AC2 | |
| 14) | chain | A |
| residue | 93 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE GOL A 202 | |
| source | : AC2 | |
| 15) | chain | A |
| residue | 95 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GOL A 202 | |
| source | : AC2 | |
| 16) | chain | A |
| residue | 96 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE GOL A 202 | |
| source | : AC2 | |
| 17) | chain | A |
| residue | 97 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE GOL A 202 | |
| source | : AC2 | |
| 18) | chain | A |
| residue | 13 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE GOL B 201 | |
| source | : AC3 | |
| 19) | chain | A |
| residue | 104 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE GOL B 201 | |
| source | : AC3 | |
| 20) | chain | A |
| residue | 128 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE GOL B 201 | |
| source | : AC3 | |
| 21) | chain | A |
| residue | 129 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE GOL B 201 | |
| source | : AC3 | |
| 22) | chain | A |
| residue | 130 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE GOL B 201 | |
| source | : AC3 | |
| 23) | chain | B |
| residue | 11 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE GOL B 201 | |
| source | : AC3 | |
| 24) | chain | B |
| residue | 107 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE GOL B 201 | |
| source | : AC3 | |
| 25) | chain | B |
| residue | 109 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE GOL B 201 | |
| source | : AC3 | |
| 26) | chain | B |
| residue | 56-66 | |
| type | prosite | |
| sequence | IAHGMLTMGIG | |
| description | THIOL_PROTEASE_HIS Eukaryotic thiol (cysteine) proteases histidine active site. IAHGMLTMGIG | |
| source | prosite : PS00639 | |