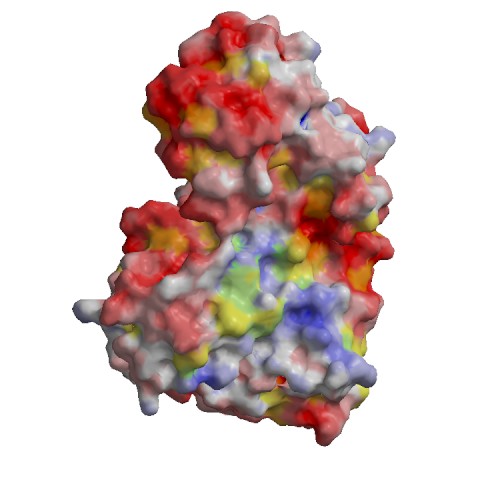
Functional site
| 1) | chain | A |
| residue | 322 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 501 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 324 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 501 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 326 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 501 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 328 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE CA A 501 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 333 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE CA A 501 | |
| source | : AC1 | |
| 6) | chain | A |
| residue | 358 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 502 | |
| source | : AC2 | |
| 7) | chain | A |
| residue | 360 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 502 | |
| source | : AC2 | |
| 8) | chain | A |
| residue | 362 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 502 | |
| source | : AC2 | |
| 9) | chain | A |
| residue | 364 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE CA A 502 | |
| source | : AC2 | |
| 10) | chain | A |
| residue | 369 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE CA A 502 | |
| source | : AC2 | |
| 11) | chain | A |
| residue | 395 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 503 | |
| source | : AC3 | |
| 12) | chain | A |
| residue | 397 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 503 | |
| source | : AC3 | |
| 13) | chain | A |
| residue | 399 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE CA A 503 | |
| source | : AC3 | |
| 14) | chain | A |
| residue | 401 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE CA A 503 | |
| source | : AC3 | |
| 15) | chain | A |
| residue | 406 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE CA A 503 | |
| source | : AC3 | |
| 16) | chain | A |
| residue | 431 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 504 | |
| source | : AC4 | |
| 17) | chain | A |
| residue | 433 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 504 | |
| source | : AC4 | |
| 18) | chain | A |
| residue | 435 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE CA A 504 | |
| source | : AC4 | |
| 19) | chain | A |
| residue | 437 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE CA A 504 | |
| source | : AC4 | |
| 20) | chain | A |
| residue | 442 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE CA A 504 | |
| source | : AC4 | |
| 21) | chain | A |
| residue | 324-336 | |
| type | prosite | |
| sequence | DGDGTITTKELGT | |
| description | EF_HAND_1 EF-hand calcium-binding domain. DKDGDGTITtkEL | |
| source | prosite : PS00018 | |
| 22) | chain | A |
| residue | 360-372 | |
| type | prosite | |
| sequence | DGDGTIDFPEFLT | |
| description | EF_HAND_1 EF-hand calcium-binding domain. DKDGDGTITtkEL | |
| source | prosite : PS00018 | |
| 23) | chain | A |
| residue | 397-409 | |
| type | prosite | |
| sequence | DGNGYISAAELRH | |
| description | EF_HAND_1 EF-hand calcium-binding domain. DKDGDGTITtkEL | |
| source | prosite : PS00018 | |
| 24) | chain | A |
| residue | 433-445 | |
| type | prosite | |
| sequence | DGDGQVNYEEFVQ | |
| description | EF_HAND_1 EF-hand calcium-binding domain. DKDGDGTITtkEL | |
| source | prosite : PS00018 | |
| 25) | chain | A |
| residue | 225 | |
| type | MOD_RES | |
| sequence | V | |
| description | (Z)-2,3-didehydrotyrosine => ECO:0000269|PubMed:8448132 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 26) | chain | A |
| residue | 226 | |
| type | CROSSLNK | |
| sequence | Q | |
| description | 5-imidazolinone (Ser-Gly) => ECO:0000269|PubMed:8448132 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 27) | chain | A |
| residue | 222 | |
| type | CROSSLNK | |
| sequence | X | |
| description | 5-imidazolinone (Ser-Gly) => ECO:0000269|PubMed:8448132 | |
| source | Swiss-Prot : SWS_FT_FI2 | |