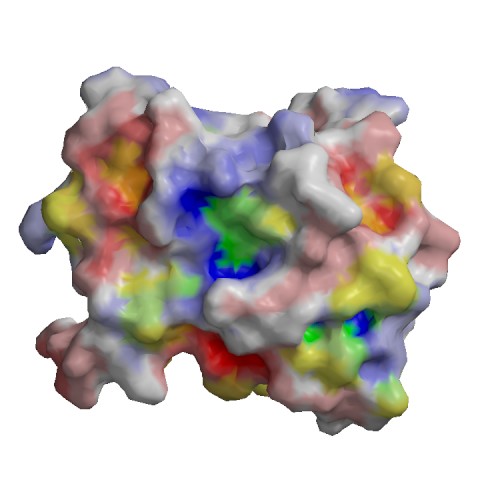
Functional site
| 1) | chain | A |
| residue | 191 | |
| type | SITE | |
| sequence | F | |
| description | Involved in dimerization of the capsid protein => ECO:0000269|PubMed:25100849 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 2) | chain | A |
| residue | 192 | |
| type | SITE | |
| sequence | Y | |
| description | Involved in dimerization of the capsid protein => ECO:0000269|PubMed:25100849 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 3) | chain | A |
| residue | 225 | |
| type | SITE | |
| sequence | N | |
| description | Involved in dimerization of the capsid protein => ECO:0000269|PubMed:25100849 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 4) | chain | A |
| residue | 267 | |
| type | SITE | |
| sequence | W | |
| description | Cleavage; by autolysis => ECO:0000250|UniProtKB:P03315 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 5) | chain | A |
| residue | 144 | |
| type | ACT_SITE | |
| sequence | H | |
| description | Charge relay system => ECO:0000255|PROSITE-ProRule:PRU01027 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 6) | chain | A |
| residue | 166 | |
| type | ACT_SITE | |
| sequence | D | |
| description | Charge relay system => ECO:0000255|PROSITE-ProRule:PRU01027 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 7) | chain | A |
| residue | 218 | |
| type | ACT_SITE | |
| sequence | S | |
| description | Charge relay system => ECO:0000255|PROSITE-ProRule:PRU01027 | |
| source | Swiss-Prot : SWS_FT_FI1 | |