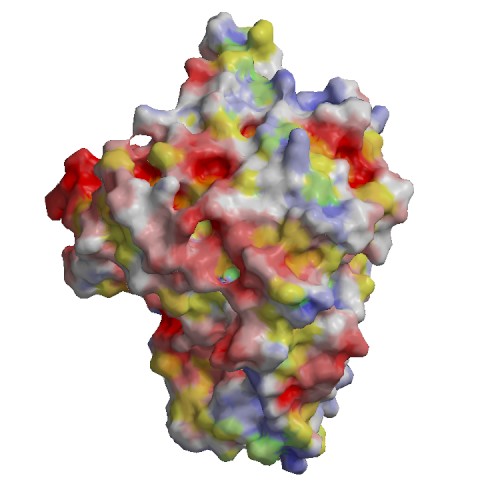
Functional site
| 1) | chain | A |
| residue | 272 | |
| type | ACT_SITE | |
| sequence | E | |
| description | Charge relay system => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 2) | chain | A |
| residue | 276 | |
| type | ACT_SITE | |
| sequence | D | |
| description | Charge relay system => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 3) | chain | A |
| residue | 475 | |
| type | ACT_SITE | |
| sequence | S | |
| description | Charge relay system => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967, ECO:0000305|PubMed:11054422 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 4) | chain | A |
| residue | 517 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 5) | chain | A |
| residue | 518 | |
| type | BINDING | |
| sequence | V | |
| description | BINDING => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 6) | chain | A |
| residue | 539 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 7) | chain | A |
| residue | 541 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 8) | chain | A |
| residue | 543 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 9) | chain | A |
| residue | 210 | |
| type | CARBOHYD | |
| sequence | N | |
| description | N-linked (GlcNAc...) asparagine => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967, ECO:0000269|PubMed:19159218 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 10) | chain | A |
| residue | 313 | |
| type | CARBOHYD | |
| sequence | N | |
| description | N-linked (GlcNAc...) asparagine => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967, ECO:0000269|PubMed:19159218 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 11) | chain | A |
| residue | 222 | |
| type | CARBOHYD | |
| sequence | N | |
| description | N-linked (GlcNAc...) asparagine => ECO:0000269|PubMed:19159218 | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 12) | chain | A |
| residue | 286 | |
| type | CARBOHYD | |
| sequence | N | |
| description | N-linked (GlcNAc...) asparagine => ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967 | |
| source | Swiss-Prot : SWS_FT_FI6 | |
| 13) | chain | A |
| residue | 443 | |
| type | CARBOHYD | |
| sequence | N | |
| description | N-linked (GlcNAc...) asparagine => ECO:0000269|PubMed:12754519, ECO:0000269|PubMed:19038966, ECO:0000269|PubMed:19038967, ECO:0000269|PubMed:19159218 | |
| source | Swiss-Prot : SWS_FT_FI7 | |