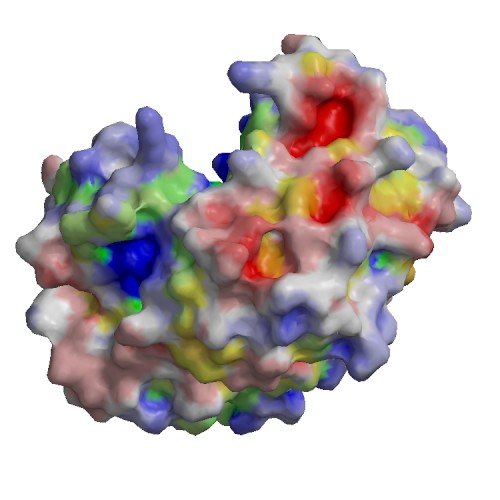
Functional site
| 1) | chain | A |
| residue | 55 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 56 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 57 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 63 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 85 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 6) | chain | A |
| residue | 90 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 7) | chain | A |
| residue | 118 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 8) | chain | A |
| residue | 188 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 9) | chain | A |
| residue | 189 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 10) | chain | A |
| residue | 214 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 11) | chain | A |
| residue | 243 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 12) | chain | A |
| residue | 244 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 13) | chain | A |
| residue | 245 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 14) | chain | A |
| residue | 246 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 15) | chain | A |
| residue | 247 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 16) | chain | A |
| residue | 248 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 17) | chain | A |
| residue | 249 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 18) | chain | A |
| residue | 252 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 19) | chain | A |
| residue | 254 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 20) | chain | A |
| residue | 264 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 21) | chain | A |
| residue | 321 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 22) | chain | A |
| residue | 322 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 23) | chain | A |
| residue | 323 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 24) | chain | A |
| residue | 325 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 25) | chain | A |
| residue | 329 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 26) | chain | A |
| residue | 332 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 27) | chain | A |
| residue | 333 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE NDP A1001 | |
| source | : AC1 | |
| 28) | chain | A |
| residue | 57 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE PDN A 501 | |
| source | : AC2 | |
| 29) | chain | A |
| residue | 89 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE PDN A 501 | |
| source | : AC2 | |
| 30) | chain | A |
| residue | 90 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE PDN A 501 | |
| source | : AC2 | |
| 31) | chain | A |
| residue | 118 | |
| type | ||
| sequence | K | |
| description | BINDING SITE FOR RESIDUE PDN A 501 | |
| source | : AC2 | |
| 32) | chain | A |
| residue | 121 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE PDN A 501 | |
| source | : AC2 | |
| 33) | chain | A |
| residue | 158 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE PDN A 501 | |
| source | : AC2 | |
| 34) | chain | A |
| residue | 189 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE PDN A 501 | |
| source | : AC2 | |
| 35) | chain | A |
| residue | 90 | |
| type | ACT_SITE | |
| sequence | Y | |
| description | Proton donor/acceptor => ECO:0000305|PubMed:18672894 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 36) | chain | A |
| residue | 158 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:4JTA | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 37) | chain | A |
| residue | 262 | |
| type | BINDING | |
| sequence | Y | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI8 | |
| 38) | chain | A |
| residue | 323 | |
| type | BINDING | |
| sequence | G | |
| description | BINDING => ECO:0000269|PubMed:23705070, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI9 | |
| 39) | chain | A |
| residue | 332 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI10 | |
| 40) | chain | A |
| residue | 112 | |
| type | MOD_RES | |
| sequence | S | |
| description | Phosphoserine => ECO:0000269|PubMed:21357749 | |
| source | Swiss-Prot : SWS_FT_FI11 | |
| 41) | chain | A |
| residue | 124 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-acetyllysine => ECO:0000250|UniProtKB:Q13303 | |
| source | Swiss-Prot : SWS_FT_FI12 | |
| 42) | chain | A |
| residue | 56 | |
| type | BINDING | |
| sequence | T | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 43) | chain | A |
| residue | 254 | |
| type | BINDING | |
| sequence | K | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 44) | chain | A |
| residue | 264 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 45) | chain | A |
| residue | 325 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 46) | chain | A |
| residue | 329 | |
| type | BINDING | |
| sequence | Q | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 47) | chain | A |
| residue | 333 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 48) | chain | A |
| residue | 57 | |
| type | BINDING | |
| sequence | W | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 49) | chain | A |
| residue | 63 | |
| type | BINDING | |
| sequence | Q | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 50) | chain | A |
| residue | 85 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 51) | chain | A |
| residue | 188 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 52) | chain | A |
| residue | 214 | |
| type | BINDING | |
| sequence | Q | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 53) | chain | A |
| residue | 243 | |
| type | BINDING | |
| sequence | W | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 54) | chain | A |
| residue | 244 | |
| type | BINDING | |
| sequence | S | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 55) | chain | A |
| residue | 246 | |
| type | BINDING | |
| sequence | L | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 56) | chain | A |
| residue | 189 | |
| type | BINDING | |
| sequence | R | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 57) | chain | A |
| residue | 245 | |
| type | BINDING | |
| sequence | P | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTC, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI5 | |
| 58) | chain | A |
| residue | 247 | |
| type | BINDING | |
| sequence | A | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:4JTA | |
| source | Swiss-Prot : SWS_FT_FI6 | |
| 59) | chain | A |
| residue | 248 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:10399921, ECO:0000269|PubMed:10884227, ECO:0000269|PubMed:16002581, ECO:0000269|PubMed:18004376, ECO:0000269|PubMed:18806782, ECO:0000269|PubMed:20360102, ECO:0000269|PubMed:20534430, ECO:0000269|PubMed:23705070, ECO:0007744|PDB:1EXB, ECO:0007744|PDB:1QRQ, ECO:0007744|PDB:2A79, ECO:0007744|PDB:2R9R, ECO:0007744|PDB:3EAU, ECO:0007744|PDB:3EB3, ECO:0007744|PDB:3EB4, ECO:0007744|PDB:3LNM, ECO:0007744|PDB:3LUT, ECO:0007744|PDB:4JTA, ECO:0007744|PDB:4JTD | |
| source | Swiss-Prot : SWS_FT_FI7 | |