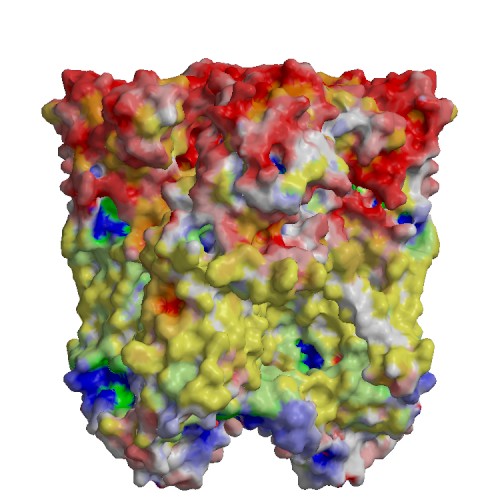
Functional site
| 1) | chain | A |
| residue | 58 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 62 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 63 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 66 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 94 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 6) | chain | A |
| residue | 97 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 7) | chain | A |
| residue | 104 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 8) | chain | A |
| residue | 142 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 9) | chain | A |
| residue | 146 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 10) | chain | A |
| residue | 149 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 11) | chain | A |
| residue | 150 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 12) | chain | A |
| residue | 198 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 13) | chain | A |
| residue | 202 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE HEM A 500 | |
| source | : AC1 | |
| 14) | chain | A |
| residue | 45 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 15) | chain | A |
| residue | 48 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 16) | chain | A |
| residue | 51 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 17) | chain | A |
| residue | 52 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 18) | chain | A |
| residue | 111 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 19) | chain | A |
| residue | 114 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 20) | chain | A |
| residue | 120 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 21) | chain | A |
| residue | 125 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 22) | chain | A |
| residue | 129 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 23) | chain | A |
| residue | 132 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 24) | chain | A |
| residue | 133 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 25) | chain | A |
| residue | 135 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 26) | chain | A |
| residue | 212 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 27) | chain | A |
| residue | 216 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 28) | chain | A |
| residue | 220 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 29) | chain | A |
| residue | 221 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 30) | chain | A |
| residue | 222 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE HEM A 501 | |
| source | : AC2 | |
| 31) | chain | A |
| residue | 137 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 32) | chain | A |
| residue | 154 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 33) | chain | A |
| residue | 158 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 34) | chain | A |
| residue | 161 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 35) | chain | A |
| residue | 194 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 36) | chain | A |
| residue | 292 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 37) | chain | A |
| residue | 295 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 38) | chain | A |
| residue | 298 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 39) | chain | A |
| residue | 302 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 40) | chain | A |
| residue | 336 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 41) | chain | A |
| residue | 337 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 42) | chain | F |
| residue | 154 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 43) | chain | F |
| residue | 155 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE SMA A 502 | |
| source | : AC3 | |
| 44) | chain | B |
| residue | 81 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 45) | chain | B |
| residue | 82 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 46) | chain | B |
| residue | 85 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 47) | chain | B |
| residue | 86 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 48) | chain | B |
| residue | 145 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 49) | chain | B |
| residue | 154 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 50) | chain | B |
| residue | 177 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 51) | chain | B |
| residue | 182 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 52) | chain | B |
| residue | 203 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 53) | chain | B |
| residue | 209 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 54) | chain | B |
| residue | 210 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE HEC B 500 | |
| source | : AC4 | |
| 55) | chain | C |
| residue | 132 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE FES C 500 | |
| source | : AC5 | |
| 56) | chain | C |
| residue | 134 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE FES C 500 | |
| source | : AC5 | |
| 57) | chain | C |
| residue | 135 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE FES C 500 | |
| source | : AC5 | |
| 58) | chain | C |
| residue | 152 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE FES C 500 | |
| source | : AC5 | |
| 59) | chain | C |
| residue | 155 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE FES C 500 | |
| source | : AC5 | |
| 60) | chain | C |
| residue | 157 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FES C 500 | |
| source | : AC5 | |
| 61) | chain | D |
| residue | 58 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 62) | chain | D |
| residue | 62 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 63) | chain | D |
| residue | 63 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 64) | chain | D |
| residue | 66 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 65) | chain | D |
| residue | 94 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 66) | chain | D |
| residue | 97 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 67) | chain | D |
| residue | 98 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 68) | chain | D |
| residue | 104 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 69) | chain | D |
| residue | 146 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 70) | chain | D |
| residue | 149 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 71) | chain | D |
| residue | 150 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 72) | chain | D |
| residue | 198 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 73) | chain | D |
| residue | 202 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE HEM D 500 | |
| source | : AC6 | |
| 74) | chain | D |
| residue | 45 | |
| type | ||
| sequence | W | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 75) | chain | D |
| residue | 48 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 76) | chain | D |
| residue | 51 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 77) | chain | D |
| residue | 52 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 78) | chain | D |
| residue | 111 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 79) | chain | D |
| residue | 114 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 80) | chain | D |
| residue | 120 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 81) | chain | D |
| residue | 125 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 82) | chain | D |
| residue | 132 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 83) | chain | D |
| residue | 133 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 84) | chain | D |
| residue | 135 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 85) | chain | D |
| residue | 209 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 86) | chain | D |
| residue | 212 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 87) | chain | D |
| residue | 216 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 88) | chain | D |
| residue | 220 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 89) | chain | D |
| residue | 221 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 90) | chain | D |
| residue | 222 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE HEM D 501 | |
| source | : AC7 | |
| 91) | chain | C |
| residue | 154 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 92) | chain | C |
| residue | 155 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 93) | chain | D |
| residue | 137 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 94) | chain | D |
| residue | 140 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 95) | chain | D |
| residue | 161 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 96) | chain | D |
| residue | 292 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 97) | chain | D |
| residue | 294 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 98) | chain | D |
| residue | 295 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 99) | chain | D |
| residue | 298 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 100) | chain | D |
| residue | 302 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 101) | chain | D |
| residue | 336 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 102) | chain | D |
| residue | 337 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 103) | chain | D |
| residue | 340 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE SMA D 502 | |
| source | : AC8 | |
| 104) | chain | E |
| residue | 81 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 105) | chain | E |
| residue | 82 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 106) | chain | E |
| residue | 85 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 107) | chain | E |
| residue | 86 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 108) | chain | E |
| residue | 154 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 109) | chain | E |
| residue | 177 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 110) | chain | E |
| residue | 203 | |
| type | ||
| sequence | F | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 111) | chain | E |
| residue | 209 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 112) | chain | E |
| residue | 210 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 113) | chain | E |
| residue | 213 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE HEC E 500 | |
| source | : AC9 | |
| 114) | chain | F |
| residue | 132 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE FES F 500 | |
| source | : BC1 | |
| 115) | chain | F |
| residue | 134 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE FES F 500 | |
| source | : BC1 | |
| 116) | chain | F |
| residue | 135 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE FES F 500 | |
| source | : BC1 | |
| 117) | chain | F |
| residue | 152 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE FES F 500 | |
| source | : BC1 | |
| 118) | chain | F |
| residue | 155 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE FES F 500 | |
| source | : BC1 | |
| 119) | chain | F |
| residue | 157 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE FES F 500 | |
| source | : BC1 | |
| 120) | chain | C |
| residue | 18-39 | |
| type | TRANSMEM | |
| sequence | FLYYATAGAGTVAAGAAAWTLV | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 121) | chain | F |
| residue | 18-39 | |
| type | TRANSMEM | |
| sequence | FLYYATAGAGTVAAGAAAWTLV | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 122) | chain | A |
| residue | 129-149 | |
| type | TRANSMEM | |
| sequence | WIVGMLIYLMMMGTAFMGYVL | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 123) | chain | A |
| residue | 156-176 | |
| type | TRANSMEM | |
| sequence | FWGATVITGLFGAIPGVGEAI | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 124) | chain | A |
| residue | 194-214 | |
| type | TRANSMEM | |
| sequence | FFSLHYLLPFVIAALVVVHIW | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 125) | chain | A |
| residue | 253-273 | |
| type | TRANSMEM | |
| sequence | LFALAVVLVVFFAIVGFMPNY | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 126) | chain | A |
| residue | 296-315 | |
| type | TRANSMEM | |
| sequence | WYFLPFYAILRAFTADVWVV | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 127) | chain | A |
| residue | 330-350 | |
| type | TRANSMEM | |
| sequence | FFGVIAMFGAILVMALVPWLD | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 128) | chain | A |
| residue | 365-385 | |
| type | TRANSMEM | |
| sequence | WWFWLLAVDFVVLMWVGAMPA | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 129) | chain | A |
| residue | 394-414 | |
| type | TRANSMEM | |
| sequence | LAGSAYWFAYFLIILPLLGII | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 130) | chain | D |
| residue | 46-66 | |
| type | TRANSMEM | |
| sequence | IWGIVLAFCLVLQIATGIVLV | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 131) | chain | D |
| residue | 100-120 | |
| type | TRANSMEM | |
| sequence | GASLFFLAVYIHIFRGLYYGS | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 132) | chain | D |
| residue | 129-149 | |
| type | TRANSMEM | |
| sequence | WIVGMLIYLMMMGTAFMGYVL | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 133) | chain | D |
| residue | 156-176 | |
| type | TRANSMEM | |
| sequence | FWGATVITGLFGAIPGVGEAI | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 134) | chain | D |
| residue | 194-214 | |
| type | TRANSMEM | |
| sequence | FFSLHYLLPFVIAALVVVHIW | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 135) | chain | D |
| residue | 253-273 | |
| type | TRANSMEM | |
| sequence | LFALAVVLVVFFAIVGFMPNY | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 136) | chain | D |
| residue | 296-315 | |
| type | TRANSMEM | |
| sequence | WYFLPFYAILRAFTADVWVV | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 137) | chain | D |
| residue | 330-350 | |
| type | TRANSMEM | |
| sequence | FFGVIAMFGAILVMALVPWLD | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 138) | chain | D |
| residue | 365-385 | |
| type | TRANSMEM | |
| sequence | WWFWLLAVDFVVLMWVGAMPA | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 139) | chain | D |
| residue | 394-414 | |
| type | TRANSMEM | |
| sequence | LAGSAYWFAYFLIILPLLGII | |
| description | Helical => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 140) | chain | C |
| residue | 152 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00628 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 141) | chain | F |
| residue | 132 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00628 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 142) | chain | F |
| residue | 134 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00628 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 143) | chain | F |
| residue | 152 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00628 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 144) | chain | F |
| residue | 155 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00628 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 145) | chain | C |
| residue | 155 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00628 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 146) | chain | C |
| residue | 132 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00628 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 147) | chain | C |
| residue | 134 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00628 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 148) | chain | B |
| residue | 86 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 149) | chain | E |
| residue | 86 | |
| type | BINDING | |
| sequence | H | |
| description | axial binding residue | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 150) | chain | B |
| residue | 210 | |
| type | BINDING | |
| sequence | M | |
| description | axial binding residue => ECO:0000255|PROSITE-ProRule:PRU00433 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 151) | chain | E |
| residue | 210 | |
| type | BINDING | |
| sequence | M | |
| description | axial binding residue => ECO:0000255|PROSITE-ProRule:PRU00433 | |
| source | Swiss-Prot : SWS_FT_FI4 | |