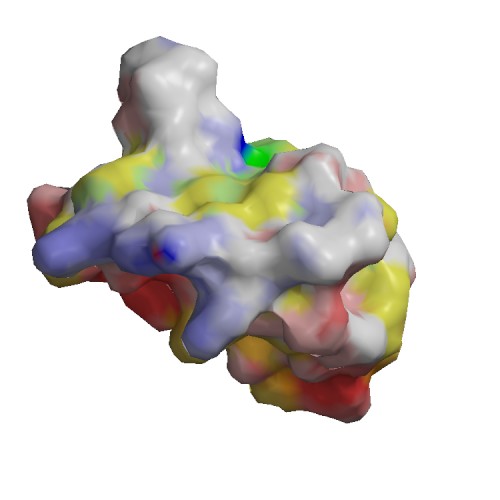
Functional site
| 1) | chain | A |
| residue | 244 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE NA A 300 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 199 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE DTT A 1245 | |
| source | : AC2 | |
| 3) | chain | A |
| residue | 202 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE DTT A 1245 | |
| source | : AC2 | |
| 4) | chain | A |
| residue | 219 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE DTT A 1245 | |
| source | : AC2 | |
| 5) | chain | A |
| residue | 199 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 1246 | |
| source | : AC4 | |
| 6) | chain | A |
| residue | 201 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 1246 | |
| source | : AC4 | |
| 7) | chain | A |
| residue | 223 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN A 1246 | |
| source | : AC4 | |
| 8) | chain | A |
| residue | 226 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 1246 | |
| source | : AC4 | |
| 9) | chain | A |
| residue | 212 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 1247 | |
| source | : AC7 | |
| 10) | chain | A |
| residue | 217 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 1247 | |
| source | : AC7 | |
| 11) | chain | A |
| residue | 239 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 1247 | |
| source | : AC7 | |
| 12) | chain | A |
| residue | 242 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 1247 | |
| source | : AC7 | |
| 13) | chain | A |
| residue | 199-242 | |
| type | prosite | |
| sequence |
CLCHQVSYGEMIGCDNPDCSIEWFHFACVGLTTKPRGKWF CPRC |
|
| description | ZF_PHD_1 Zinc finger PHD-type signature. Cl.Chqvsygem.....................................IgCdnpdCsiewFHfaCvglttkprgk...................................WfCprC | |
| source | prosite : PS01359 | |
| 14) | chain | A |
| residue | 212 | |
| type | MOD_RES | |
| sequence | C | |
| description | Phosphothreonine; by HASPIN and VRK1 => ECO:0000269|PubMed:15681610, ECO:0000269|PubMed:16185088, ECO:0000269|PubMed:31527692 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 15) | chain | A |
| residue | 217 | |
| type | MOD_RES | |
| sequence | C | |
| description | Phosphothreonine; by HASPIN and VRK1 => ECO:0000269|PubMed:15681610, ECO:0000269|PubMed:16185088, ECO:0000269|PubMed:31527692 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 16) | chain | A |
| residue | 223 | |
| type | MOD_RES | |
| sequence | H | |
| description | Phosphothreonine; by HASPIN and VRK1 => ECO:0000269|PubMed:15681610, ECO:0000269|PubMed:16185088, ECO:0000269|PubMed:31527692 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 17) | chain | A |
| residue | 226 | |
| type | MOD_RES | |
| sequence | C | |
| description | Phosphothreonine; by HASPIN and VRK1 => ECO:0000269|PubMed:15681610, ECO:0000269|PubMed:16185088, ECO:0000269|PubMed:31527692 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 18) | chain | A |
| residue | 239 | |
| type | MOD_RES | |
| sequence | C | |
| description | Phosphothreonine; by HASPIN and VRK1 => ECO:0000269|PubMed:15681610, ECO:0000269|PubMed:16185088, ECO:0000269|PubMed:31527692 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 19) | chain | A |
| residue | 242 | |
| type | MOD_RES | |
| sequence | C | |
| description | Phosphothreonine; by HASPIN and VRK1 => ECO:0000269|PubMed:15681610, ECO:0000269|PubMed:16185088, ECO:0000269|PubMed:31527692 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 20) | chain | A |
| residue | 213 | |
| type | MOD_RES | |
| sequence | D | |
| description | N6-methyllysine; alternate => ECO:0000269|PubMed:16267050, ECO:0000269|PubMed:16457588, ECO:0000269|PubMed:17194708 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 21) | chain | A |
| residue | 221 | |
| type | MOD_RES | |
| sequence | W | |
| description | N6-methyllysine; alternate => ECO:0000269|PubMed:16267050, ECO:0000269|PubMed:16457588, ECO:0000269|PubMed:17194708 | |
| source | Swiss-Prot : SWS_FT_FI3 | |