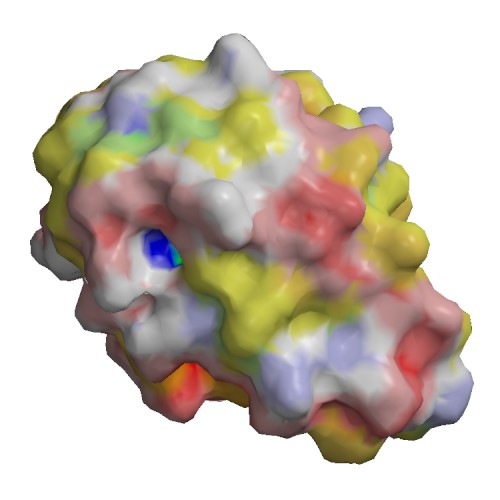
Functional site
| 1) | chain | E |
| residue | 45 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE F3S E 104 | |
| source | : BC4 | |
| 2) | chain | E |
| residue | 46 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE F3S E 104 | |
| source | : BC4 | |
| 3) | chain | E |
| residue | 47 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE F3S E 104 | |
| source | : BC4 | |
| 4) | chain | E |
| residue | 48 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE F3S E 104 | |
| source | : BC4 | |
| 5) | chain | E |
| residue | 49 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE F3S E 104 | |
| source | : BC4 | |
| 6) | chain | E |
| residue | 50 | |
| type | ||
| sequence | S | |
| description | BINDING SITE FOR RESIDUE F3S E 104 | |
| source | : BC4 | |
| 7) | chain | E |
| residue | 51 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE F3S E 104 | |
| source | : BC4 | |
| 8) | chain | E |
| residue | 93 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE F3S E 104 | |
| source | : BC4 | |
| 9) | chain | E |
| residue | 55 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE SF4 E 105 | |
| source | : BC5 | |
| 10) | chain | E |
| residue | 59 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE SF4 E 105 | |
| source | : BC5 | |
| 11) | chain | E |
| residue | 83 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE SF4 E 105 | |
| source | : BC5 | |
| 12) | chain | E |
| residue | 84 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE SF4 E 105 | |
| source | : BC5 | |
| 13) | chain | E |
| residue | 86 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE SF4 E 105 | |
| source | : BC5 | |
| 14) | chain | E |
| residue | 87 | |
| type | ||
| sequence | M | |
| description | BINDING SITE FOR RESIDUE SF4 E 105 | |
| source | : BC5 | |
| 15) | chain | E |
| residue | 89 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE SF4 E 105 | |
| source | : BC5 | |
| 16) | chain | E |
| residue | 16 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN E 106 | |
| source | : BC6 | |
| 17) | chain | E |
| residue | 19 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN E 106 | |
| source | : BC6 | |
| 18) | chain | E |
| residue | 34 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN E 106 | |
| source | : BC6 | |
| 19) | chain | E |
| residue | 76 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE ZN E 106 | |
| source | : BC6 | |
| 20) | chain | E |
| residue | 16 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 21) | chain | E |
| residue | 19 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 22) | chain | E |
| residue | 34 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 23) | chain | E |
| residue | 45 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 24) | chain | E |
| residue | 46 | |
| type | BINDING | |
| sequence | I | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 25) | chain | E |
| residue | 51 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 26) | chain | E |
| residue | 55 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 27) | chain | E |
| residue | 76 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 28) | chain | E |
| residue | 83 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 29) | chain | E |
| residue | 86 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 30) | chain | E |
| residue | 89 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 31) | chain | E |
| residue | 93 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:18258200, ECO:0007744|PDB:2VKR | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 32) | chain | E |
| residue | 101 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-methyllysine; partial => ECO:0000269|Ref.1 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 33) | chain | E |
| residue | 29 | |
| type | MOD_RES | |
| sequence | K | |
| description | N6-methyllysine; partial => ECO:0000269|Ref.1 | |
| source | Swiss-Prot : SWS_FT_FI2 | |