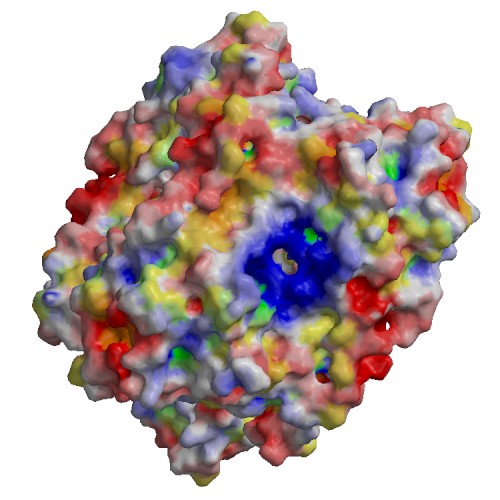
Functional site
| 1) | chain | A |
| residue | 625 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 A 801 | |
| source | : AC1 | |
| 2) | chain | A |
| residue | 649 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 A 801 | |
| source | : AC1 | |
| 3) | chain | A |
| residue | 657 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE SO4 A 801 | |
| source | : AC1 | |
| 4) | chain | A |
| residue | 660 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE SO4 A 801 | |
| source | : AC1 | |
| 5) | chain | A |
| residue | 487 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE ACP A 802 | |
| source | : AC2 | |
| 6) | chain | A |
| residue | 515 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACP A 802 | |
| source | : AC2 | |
| 7) | chain | A |
| residue | 564 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE ACP A 802 | |
| source | : AC2 | |
| 8) | chain | A |
| residue | 565 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE ACP A 802 | |
| source | : AC2 | |
| 9) | chain | A |
| residue | 567 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACP A 802 | |
| source | : AC2 | |
| 10) | chain | A |
| residue | 571 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE ACP A 802 | |
| source | : AC2 | |
| 11) | chain | A |
| residue | 633 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE ACP A 802 | |
| source | : AC2 | |
| 12) | chain | B |
| residue | 625 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 B 8001 | |
| source | : AC3 | |
| 13) | chain | B |
| residue | 649 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 B 8001 | |
| source | : AC3 | |
| 14) | chain | B |
| residue | 657 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE SO4 B 8001 | |
| source | : AC3 | |
| 15) | chain | B |
| residue | 664 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 B 8001 | |
| source | : AC3 | |
| 16) | chain | B |
| residue | 487 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 17) | chain | B |
| residue | 495 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 18) | chain | B |
| residue | 515 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 19) | chain | B |
| residue | 564 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 20) | chain | B |
| residue | 565 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 21) | chain | B |
| residue | 566 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 22) | chain | B |
| residue | 567 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 23) | chain | B |
| residue | 571 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 24) | chain | B |
| residue | 630 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 25) | chain | B |
| residue | 633 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 26) | chain | B |
| residue | 644 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE ACP B 8002 | |
| source | : AC4 | |
| 27) | chain | C |
| residue | 625 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 C 801 | |
| source | : AC5 | |
| 28) | chain | C |
| residue | 649 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 C 801 | |
| source | : AC5 | |
| 29) | chain | C |
| residue | 657 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE SO4 C 801 | |
| source | : AC5 | |
| 30) | chain | C |
| residue | 664 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 C 801 | |
| source | : AC5 | |
| 31) | chain | C |
| residue | 487 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE ACP C 802 | |
| source | : AC6 | |
| 32) | chain | C |
| residue | 515 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACP C 802 | |
| source | : AC6 | |
| 33) | chain | C |
| residue | 564 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE ACP C 802 | |
| source | : AC6 | |
| 34) | chain | C |
| residue | 565 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE ACP C 802 | |
| source | : AC6 | |
| 35) | chain | C |
| residue | 566 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE ACP C 802 | |
| source | : AC6 | |
| 36) | chain | C |
| residue | 567 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACP C 802 | |
| source | : AC6 | |
| 37) | chain | C |
| residue | 571 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE ACP C 802 | |
| source | : AC6 | |
| 38) | chain | C |
| residue | 633 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE ACP C 802 | |
| source | : AC6 | |
| 39) | chain | D |
| residue | 625 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 D 801 | |
| source | : AC7 | |
| 40) | chain | D |
| residue | 649 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 D 801 | |
| source | : AC7 | |
| 41) | chain | D |
| residue | 657 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE SO4 D 801 | |
| source | : AC7 | |
| 42) | chain | D |
| residue | 660 | |
| type | ||
| sequence | T | |
| description | BINDING SITE FOR RESIDUE SO4 D 801 | |
| source | : AC7 | |
| 43) | chain | D |
| residue | 664 | |
| type | ||
| sequence | R | |
| description | BINDING SITE FOR RESIDUE SO4 D 801 | |
| source | : AC7 | |
| 44) | chain | D |
| residue | 487 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE ACP D 802 | |
| source | : AC8 | |
| 45) | chain | D |
| residue | 515 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACP D 802 | |
| source | : AC8 | |
| 46) | chain | D |
| residue | 564 | |
| type | ||
| sequence | V | |
| description | BINDING SITE FOR RESIDUE ACP D 802 | |
| source | : AC8 | |
| 47) | chain | D |
| residue | 565 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE ACP D 802 | |
| source | : AC8 | |
| 48) | chain | D |
| residue | 566 | |
| type | ||
| sequence | Y | |
| description | BINDING SITE FOR RESIDUE ACP D 802 | |
| source | : AC8 | |
| 49) | chain | D |
| residue | 567 | |
| type | ||
| sequence | A | |
| description | BINDING SITE FOR RESIDUE ACP D 802 | |
| source | : AC8 | |
| 50) | chain | D |
| residue | 571 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE ACP D 802 | |
| source | : AC8 | |
| 51) | chain | D |
| residue | 633 | |
| type | ||
| sequence | L | |
| description | BINDING SITE FOR RESIDUE ACP D 802 | |
| source | : AC8 | |
| 52) | chain | A |
| residue | 626 | |
| type | ACT_SITE | |
| sequence | D | |
| description | Proton acceptor => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000255|PROSITE-ProRule:PRU10028, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 53) | chain | B |
| residue | 626 | |
| type | ACT_SITE | |
| sequence | D | |
| description | Proton acceptor => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000255|PROSITE-ProRule:PRU10028, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 54) | chain | C |
| residue | 626 | |
| type | ACT_SITE | |
| sequence | D | |
| description | Proton acceptor => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000255|PROSITE-ProRule:PRU10028, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 55) | chain | D |
| residue | 626 | |
| type | ACT_SITE | |
| sequence | D | |
| description | Proton acceptor => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000255|PROSITE-ProRule:PRU10028, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 56) | chain | A |
| residue | 487 | |
| type | BINDING | |
| sequence | L | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 57) | chain | A |
| residue | 517 | |
| type | BINDING | |
| sequence | K | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 58) | chain | A |
| residue | 565 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 59) | chain | A |
| residue | 571 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 60) | chain | C |
| residue | 517 | |
| type | BINDING | |
| sequence | K | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 61) | chain | C |
| residue | 565 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 62) | chain | C |
| residue | 571 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 63) | chain | D |
| residue | 487 | |
| type | BINDING | |
| sequence | L | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 64) | chain | D |
| residue | 517 | |
| type | BINDING | |
| sequence | K | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 65) | chain | D |
| residue | 565 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 66) | chain | D |
| residue | 571 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 67) | chain | B |
| residue | 487 | |
| type | BINDING | |
| sequence | L | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 68) | chain | B |
| residue | 517 | |
| type | BINDING | |
| sequence | K | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 69) | chain | B |
| residue | 565 | |
| type | BINDING | |
| sequence | E | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 70) | chain | B |
| residue | 571 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 71) | chain | C |
| residue | 487 | |
| type | BINDING | |
| sequence | L | |
| description | BINDING => ECO:0000255|PROSITE-ProRule:PRU00159, ECO:0000269|PubMed:19060208 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 72) | chain | B |
| residue | 466 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 73) | chain | C |
| residue | 466 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 74) | chain | A |
| residue | 656 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:17803937, ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 75) | chain | A |
| residue | 657 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:17803937, ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 76) | chain | B |
| residue | 656 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:17803937, ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 77) | chain | B |
| residue | 657 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:17803937, ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 78) | chain | C |
| residue | 656 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:17803937, ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 79) | chain | D |
| residue | 656 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:17803937, ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 80) | chain | D |
| residue | 657 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:17803937, ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 81) | chain | C |
| residue | 657 | |
| type | MOD_RES | |
| sequence | Y | |
| description | Phosphotyrosine; by autocatalysis => ECO:0000269|PubMed:17803937, ECO:0000269|PubMed:19060208, ECO:0000269|PubMed:19410646 | |
| source | Swiss-Prot : SWS_FT_FI4 | |
| 82) | chain | A |
| residue | 487-517 | |
| type | prosite | |
| sequence | LQVVMAEAVGIDKDKPKEAVTVAVK | |
| description | PROTEIN_KINASE_ATP Protein kinases ATP-binding region signature. LGEGAFGQVVmAeavgidkdkpkeavt...VAVK | |
| source | prosite : PS00107 | |
| 83) | chain | A |
| residue | 622-634 | |
| type | prosite | |
| sequence | CIHRDLAARNVLV | |
| description | PROTEIN_KINASE_TYR Tyrosine protein kinases specific active-site signature. CIHrDLAARNVLV | |
| source | prosite : PS00109 | |