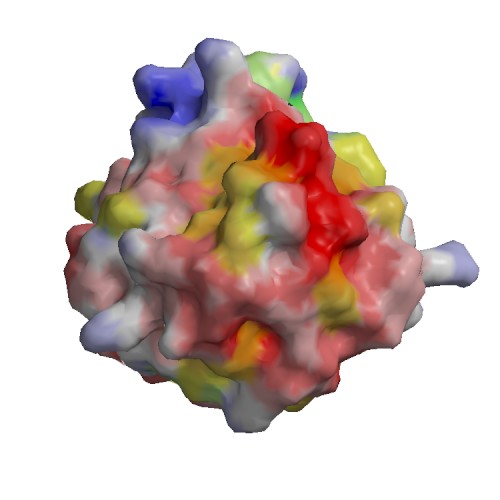
Functional site
| 1) | chain | A |
| residue | 62 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN A 401 | |
| source | : AC2 | |
| 2) | chain | A |
| residue | 91 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 401 | |
| source | : AC2 | |
| 3) | chain | A |
| residue | 94 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE ZN A 401 | |
| source | : AC2 | |
| 4) | chain | A |
| residue | 128 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE CA B 403 | |
| source | : AC3 | |
| 5) | chain | A |
| residue | 150 | |
| type | ||
| sequence | Q | |
| description | BINDING SITE FOR RESIDUE CA B 403 | |
| source | : AC3 | |
| 6) | chain | A |
| residue | 33 | |
| type | ||
| sequence | I | |
| description | BINDING SITE FOR RESIDUE HPY A 411 | |
| source | : AC5 | |
| 7) | chain | A |
| residue | 51 | |
| type | ||
| sequence | N | |
| description | BINDING SITE FOR RESIDUE HPY A 411 | |
| source | : AC5 | |
| 8) | chain | A |
| residue | 62 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE HPY A 411 | |
| source | : AC5 | |
| 9) | chain | A |
| residue | 63 | |
| type | ||
| sequence | G | |
| description | BINDING SITE FOR RESIDUE HPY A 411 | |
| source | : AC5 | |
| 10) | chain | A |
| residue | 64 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE HPY A 411 | |
| source | : AC5 | |
| 11) | chain | A |
| residue | 90 | |
| type | ||
| sequence | P | |
| description | BINDING SITE FOR RESIDUE HPY A 411 | |
| source | : AC5 | |
| 12) | chain | A |
| residue | 91 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HPY A 411 | |
| source | : AC5 | |
| 13) | chain | A |
| residue | 94 | |
| type | ||
| sequence | C | |
| description | BINDING SITE FOR RESIDUE HPY A 411 | |
| source | : AC5 | |
| 14) | chain | A |
| residue | 155 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE HPY A 411 | |
| source | : AC5 | |
| 15) | chain | A |
| residue | 62 | |
| type | catalytic | |
| sequence | H | |
| description | 636 | |
| source | MCSA : MCSA1 | |
| 16) | chain | A |
| residue | 64 | |
| type | catalytic | |
| sequence | E | |
| description | 636 | |
| source | MCSA : MCSA1 | |
| 17) | chain | A |
| residue | 89 | |
| type | catalytic | |
| sequence | S | |
| description | 636 | |
| source | MCSA : MCSA1 | |
| 18) | chain | A |
| residue | 91 | |
| type | catalytic | |
| sequence | C | |
| description | 636 | |
| source | MCSA : MCSA1 | |
| 19) | chain | A |
| residue | 94 | |
| type | catalytic | |
| sequence | C | |
| description | 636 | |
| source | MCSA : MCSA1 | |
| 20) | chain | A |
| residue | 64 | |
| type | ACT_SITE | |
| sequence | E | |
| description | Proton donor => ECO:0000269|PubMed:12637534, ECO:0007744|PDB:1UAQ | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 21) | chain | A |
| residue | 51 | |
| type | BINDING | |
| sequence | N | |
| description | BINDING => ECO:0000269|PubMed:12637534, ECO:0000269|PubMed:12906827, ECO:0007744|PDB:1P6O, ECO:0007744|PDB:1UAQ | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 22) | chain | A |
| residue | 155 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:12637534, ECO:0000269|PubMed:12906827, ECO:0007744|PDB:1P6O, ECO:0007744|PDB:1UAQ | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 23) | chain | A |
| residue | 62 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000269|PubMed:12637534, ECO:0000269|PubMed:12906827, ECO:0000269|PubMed:15879217, ECO:0007744|PDB:1OX7, ECO:0007744|PDB:1P6O, ECO:0007744|PDB:1RB7, ECO:0007744|PDB:1UAQ, ECO:0007744|PDB:1YSB, ECO:0007744|PDB:1YSD | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 24) | chain | A |
| residue | 91 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:12637534, ECO:0000269|PubMed:12906827, ECO:0000269|PubMed:15879217, ECO:0007744|PDB:1OX7, ECO:0007744|PDB:1P6O, ECO:0007744|PDB:1RB7, ECO:0007744|PDB:1UAQ, ECO:0007744|PDB:1YSB, ECO:0007744|PDB:1YSD | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 25) | chain | A |
| residue | 94 | |
| type | BINDING | |
| sequence | C | |
| description | BINDING => ECO:0000269|PubMed:12637534, ECO:0000269|PubMed:12906827, ECO:0000269|PubMed:15879217, ECO:0007744|PDB:1OX7, ECO:0007744|PDB:1P6O, ECO:0007744|PDB:1RB7, ECO:0007744|PDB:1UAQ, ECO:0007744|PDB:1YSB, ECO:0007744|PDB:1YSD | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 26) | chain | A |
| residue | 62-98 | |
| type | prosite | |
| sequence | HGEISTLENCGRLEGKVYKDTTLYTTLSPCEMCTGAI | |
| description | CYT_DCMP_DEAMINASES_1 Cytidine and deoxycytidylate deaminases zinc-binding region signature. HGEisTLencgrlegkvykdttlyttls............PCem......CtgaI | |
| source | prosite : PS00903 | |