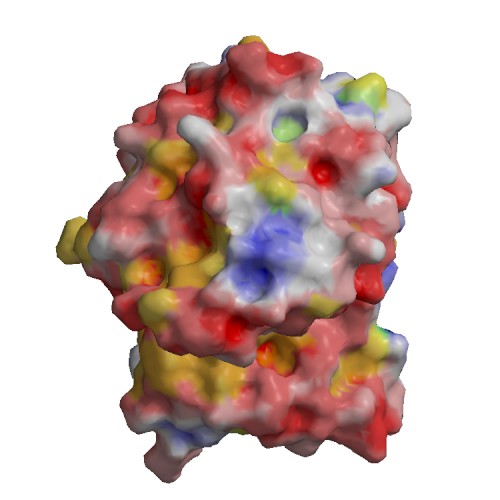
Functional site
| 1) | chain | E |
| residue | 51 | |
| type | ||
| sequence | D | |
| description | BINDING SITE FOR RESIDUE ZN E 301 | |
| source | : AC5 | |
| 2) | chain | E |
| residue | 53 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN E 301 | |
| source | : AC5 | |
| 3) | chain | E |
| residue | 94 | |
| type | ||
| sequence | H | |
| description | BINDING SITE FOR RESIDUE ZN E 301 | |
| source | : AC5 | |
| 4) | chain | E |
| residue | 129 | |
| type | ||
| sequence | E | |
| description | BINDING SITE FOR RESIDUE ZN E 301 | |
| source | : AC5 | |
| 5) | chain | E |
| residue | 122 | |
| type | ACT_SITE | |
| sequence | K | |
| description | ACT_SITE => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 6) | chain | E |
| residue | 8 | |
| type | ACT_SITE | |
| sequence | D | |
| description | ACT_SITE => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI1 | |
| 7) | chain | E |
| residue | 167 | |
| type | ACT_SITE | |
| sequence | C | |
| description | Nucleophile => ECO:0000255 | |
| source | Swiss-Prot : SWS_FT_FI2 | |
| 8) | chain | E |
| residue | 51 | |
| type | BINDING | |
| sequence | D | |
| description | BINDING => ECO:0000269|PubMed:17382284, ECO:0007744|PDB:2H0R | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 9) | chain | E |
| residue | 53 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000269|PubMed:17382284, ECO:0007744|PDB:2H0R | |
| source | Swiss-Prot : SWS_FT_FI3 | |
| 10) | chain | E |
| residue | 94 | |
| type | BINDING | |
| sequence | H | |
| description | BINDING => ECO:0000269|PubMed:17382284, ECO:0007744|PDB:2H0R | |
| source | Swiss-Prot : SWS_FT_FI3 | |